
APA Style
Bilal Rah, Menattullah W. Mohamed, Aisha S. Janeeh, Tayma M. Kurabi , Jasmin S. Abdul Salam, Mawieh Hamad. (2025). Fatty Allies: How Short-Chain Fatty Acids Turn the Tumor Microenvironment Against Cancer. Cancer Immunology Connect, 1 (Article ID: 0007). https://doi.org/10.69709/CIConnect.2025.155412MLA Style
Bilal Rah, Menattullah W. Mohamed, Aisha S. Janeeh, Tayma M. Kurabi , Jasmin S. Abdul Salam, Mawieh Hamad. "Fatty Allies: How Short-Chain Fatty Acids Turn the Tumor Microenvironment Against Cancer". Cancer Immunology Connect, vol. 1, 2025, Article ID: 0007, https://doi.org/10.69709/CIConnect.2025.155412.Chicago Style
Bilal Rah, Menattullah W. Mohamed, Aisha S. Janeeh, Tayma M. Kurabi , Jasmin S. Abdul Salam, Mawieh Hamad. 2025. "Fatty Allies: How Short-Chain Fatty Acids Turn the Tumor Microenvironment Against Cancer." Cancer Immunology Connect 1 (2025): 0007. https://doi.org/10.69709/CIConnect.2025.155412.
 ACCESS
Editorial
ACCESS
Editorial
Volume 1, Article ID: 2025.0007

Bilal Rah
BRah@sharjah.ac.ae

Menattullah W. Mohamed
U24103098@sharjah.ac.ae

Aisha S. Janeeh
U21103311@sharjah.ac.ae

Tayma M. Kurabi
U24103495@sharjah.ac.ae

Jasmin S. Abdul Salam
jsalam@sharjah.ac.ae

Mawieh Hamad
mabdelhaq@sharjah.ac.ae
1 Research Institute of Medical and Health Sciences, University of Sharjah, Sharjah-27272, United Arab Emirates
2 Department of Medical Laboratory Sciences, College of Health Sciences, University of Sharjah, Sharjah-27272, United Arab Emirates
3 College of Medicine, University of Sharjah, Sharjah-27272, United Arab Emirates
* Author to whom correspondence should be addressed
Received: 20 Apr 2025 Accepted: 15 May 2025 Published: 23 May 2025
Short-chain fatty acids (SCFAs), which are fatty acids with fewer than six carbon atoms, are primarily produced by gut microbiota through fermentation of dietary fibers. They interact with various receptors on target cells, including G-protein coupled receptors (GPCRs) and free fatty acid receptors (FFARs). These interactions induce significant epigenetic modifications, such as the inhibition of histone deacetylases (HDACs) and the regulation of several microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). SCFAs reprogram the tumor microenvironment (TME) by promoting the recruitment and effector function of cluster of differentiation (CD)8⁺ T cells and reducing regulatory T cell (Treg) activity, thereby converting the TME from an immunosuppressive to an immunoreactive state. They modulate the differentiation and activity of various immune cell subsets, including macrophages, dendritic cells (DCs), natural killer (NK) cells, neutrophils, T cells, B cells, and myeloid-derived suppressor cells (MDSCs). SCFAs also precipitate anti-growth effects in tumor cells by hampering glycolysis and enhancing fatty acid oxidation. The idea of integrating SCFAs with immunotherapy for cancer treatment has garnered considerable interest in recent years. However, more work is still needed to better understand the molecular underpinnings of the interaction between SCFAs and different cell subsets within the TME. Further investigation of issues relating to dosing, therapeutic efficacy, and safety of exogenously administered SCFAs in cancer patients, particularly immunocompromised patients, is also in order.
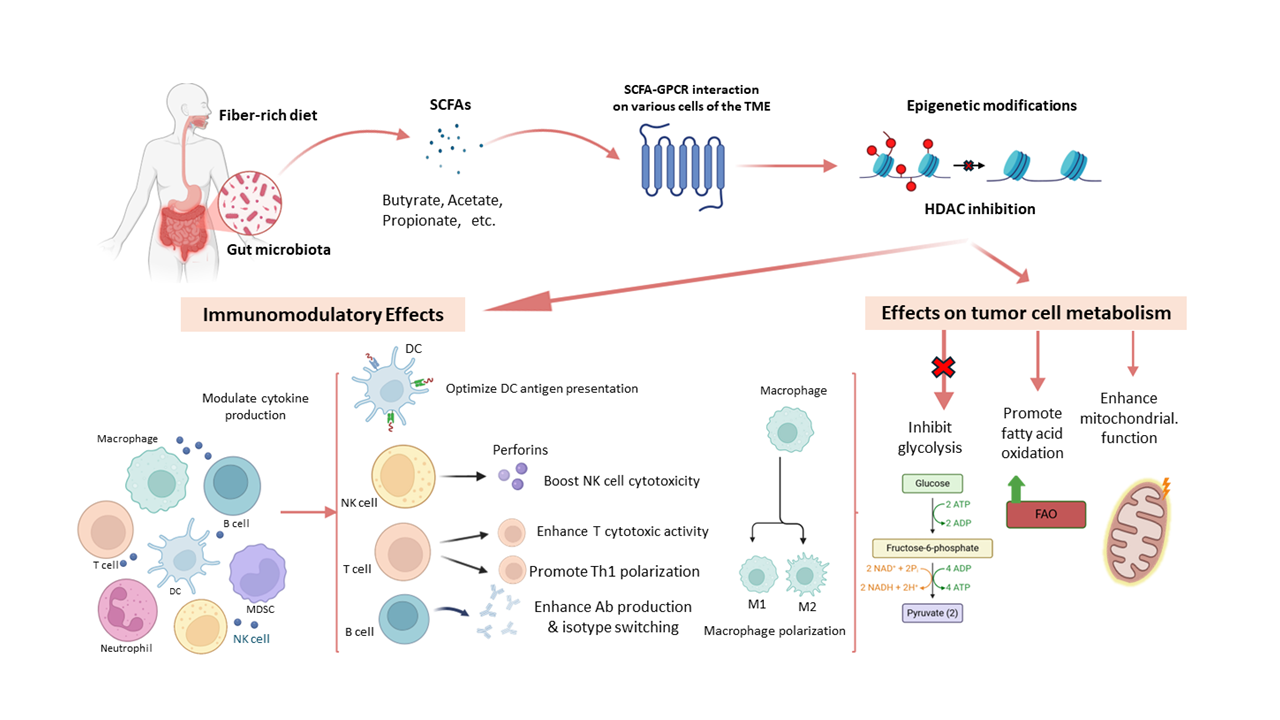
SCFAs typically contain fewer than six carbon atoms. They are produced primarily through the fermentation of dietary fibers by beneficial gut bacteria in the colon. The most common SCFAs include acetate, propionate, and butyrate. The TME is a complex system consisting of immune, stromal, and tumor cells that interact to influence cancer progression [1]. Innate and adaptive anti-tumor immune responses within the TME are key to tumor control [2]. However, tumors often evade immunity, weakening surveillance and enabling progression [3]. Mounting evidence suggests that SCFAs play a significant role in transforming the immunosuppressive TME into one that supports effective anti-tumor immunity [4]. SCFAs mediate their effects through signaling cascades transduced via several SCFA receptors, including some GPCRs and FFARs on target cells. This leads to significant epigenetic modifications in the form of methylation and the inhibition of HDACs [5]. These changes modulate several inflammatory pathways affecting macrophage polarization, NK cell activity, DC antigen presentation, and lymphocyte (T and B cells) functional differentiation within the TME [6]. SCFAs also variably influence tumor metabolism by altering glycolysis, enhancing oxidative phosphorylation, and regulating lipid metabolism depending on the type of tumor [7]. They also modulate MDSCs, key immunosuppressive cells in the TME [8]. Although SCFAs present considerable therapeutic potential as they enhance anti-tumor immunity, the risk of overwhelming the immune response and/or promoting immune evasion of tumor cells cannot be discounted [9]. More research is needed to fully understand the molecular mechanisms by which SCFAs enhance anti-tumor immunity while simultaneously suppressing tumor growth. Additionally, many aspects of the clinical application of SCFAs as adjunctive therapy in cancer patients receiving immunotherapy still need to be ironed out. This minireview commences with a general introduction to SCFAs and their anti-tumor effects and concludes with some key unanswered questions regarding their basic biology and clinical utility.
2.1. SCFAs and Innate Immunity Innate immunity is the first line of defense against tumors, detecting damage- and tumor-associated molecular patterns (DAMPs, TAMPs) to trigger inflammation and immune surveillance [10]. SCFAs modulate innate immune cells in the TME in several ways that affect tumor progression. In general terms, macrophages can polarize into an M1 (pro-inflammatory) or an M2 (anti-inflammatory) state. SCFAs such as butyrate and propionate tend to promote M1 polarization through transcriptional and epigenetic regulation [11]. They upregulate the expression of tumor necrosis factor-alpha (TNF-α), interleukin-12 (IL-12), interferon gamma (IFN-γ), nitric oxide (NO), while suppressing the expression of several M2 markers, including arginase 1 (ARG1), IL-10, and tumor growth factor-beta (TGF-β), and inhibiting HDAC from blocking tumor-promoting genes [9,12,13]. In DCs, SCFAs enhance the expression of major histocompatibility complex-II (MHC-II), CD80, and CD86, which facilitate T cell antigen receptor (TCR) and the co-stimulatory receptor CD28 engagement, thereby boosting T-cell priming [14,15]. In some contexts, SCFAs have been shown to promote the rise of tolerogenic DCs, increase IL-10 production, and reduce IL-12 production, thus contributing to immune evasion [16]. SCFAs can enhance NK cell cytotoxicity by upregulating the expression of natural killer group 2 member D (NKG2D), perforin, and granzyme B, and by improving NK’s metabolic fitness through mitochondrial biogenesis and fatty acid oxidation [17]. HDAC inhibition further amplifies the expression of NK effector genes [18]. Neutrophils are also modulated by SCFAs through various chemokines, including CXCR1 and CXCL2. For example, SCFAs enhance the capacity of neutrophils to generate reactive oxygen species (ROS) and perform phagocytosis, but whether they affect neutrophil extracellular trap (NET) formation (NETosis) remains unclear [19,20]. SCFAs modulate immune cell migration, chemokine and cytokine release, and inflammatory signaling in the TME, primarily through their ability to interact with innate receptors, including toll-like receptors (TLRs) and GPCRs (GPR41, GPR43, GPR109A) [21]. 2.2. SCFAs and Adaptive Immunity Several studies have elaborated on the capacity of SCFAs to modulate T and B lymphocytes through epigenetic regulation, metabolic reprogramming, and cytokine signaling [22]. By inhibiting HDACs and altering metabolism, SCFAs enhance CD8⁺ T cell differentiation and the production of IFN-γ, granzyme B, and perforin for cytotoxic activity, and promote the generation of adaptive memory for sustained tumor immunosurveillance [23,24]. Moreover, SCFAs promote T helper 1 (Th1) differentiation, thereby enhancing IFN-γ and IL-2 production, suppressing Th2 cytokines, eventually reducing IL-4 and IL-5, and modulating Th17 activity in a way that maintains anti-tumor immunity while limiting pathogenic inflammation [18,25,26]. Butyrate and propionate promote the expansion of forkhead box P3 (Foxp3)⁺ regulatory T (Treg) cells through histone deacetylase (HDAC) inhibition, thereby contributing to immune tolerance. However, they may contribute to the development of an immunosuppressive TME, especially when excessive accumulation of Tregs occurs [27]. While SCFA-induced Tregs may mitigate chronic inflammation associated with tumorigenesis, the fact that they can precipitate immunosuppressive effects calls for caution when considering their therapeutic utility. SCFAs enhance cytotoxic T lymphocyte (CTL) survival and function, and hence promote CTL-mediated tumor eradication through their ability to support metabolic adaptability (mitochondrial function and fatty acid oxidation), especially in nutrient-deprived TME [28]. They also reduce T cell exhaustion by decreasing the expression of programmed cell death protein 1 (PD-1), T-cell immunoglobulin and mucin-domain containing-3 (TIM-3), and lymphocyte activation gene 3 (LAG-3). Combined, these effects are believed to improve patient responsiveness to checkpoint blockade and to expand the T cell memory pool [29,30]. SCFAs also seem to modulate humoral immunity within the TME by, for example, promoting B cell proliferation and enhancing isotype (IgA/IgG ) switching [31]. So far, little is known regarding the effects of SCFAs on the differentiation or activity of TME immunosuppressive regulatory B cells (Bregs) [32]. 2.3. SCFAs and MDSCs The TME is an immunosuppressive niche that promotes tumor growth and immune evasion [33]. MDSCs, key TME components, suppress T cell activity and promote Treg expansion through several mediators, including ARG1, inducible nitric oxide synthase (iNOS), ROS, and TGF-β [34]. Targeting MDSCs is thus a promising strategy in cancer immunotherapy. Gut microbiota-derived SCFAs (acetate, propionate, and butyrate) modulate immunity through SCFA receptors (GPCRs) GPR41, GPR43, GPR109A, and through the inhibition of HDACs [35,36]. These pathways affect immune cell differentiation and function, including that of MDSCs [37]. The effects of SCFAs on MDSCs are context-dependent. For example, butyrate and propionate promote MDSC expansion and immunosuppressive activities by enhancing ARG1 and iNOS expression [38]. Conversely, SCFAs may reprogram MDSCs toward less suppressive phenotypes. Butyrate-driven HDAC inhibition and GPR109A signaling have been linked to reduced MDSC-mediated suppression and improved anti-tumor immunity [39,40]. SCFAs also shape MDSC metabolism, influencing glycolysis, oxidative phosphorylation, and fatty acid metabolism. Butyrate, in particular, promotes oxidative metabolism and mitochondrial function, potentially impairing MDSC suppressive activity [41,42]. Understanding these paradoxical effects of SCFAs on MDSC regulation may help identify novel strategies to enhance anti-tumor immunity. 2.4. SCFAs and Tumor Cell Metabolism Being metabolically adept enables cancer cells to sustain growth, resist apoptosis, and evade immunity [43]. SCFAs, as key metabolic modulators in the TME, influence glucose and lipid metabolism, mitochondrial function, and redox balance [44,45]. Generally, cancer cells favor aerobic glycolysis (Warburg effect) for adenosine triphosphate (ATP) production. SCFAs counter this by downregulating the production of the glycolytic enzymes hexokinase 2 (HK2) and pyruvate kinase M2 (PKM2) and reducing glucose uptake and lactate output [46]. SCFAs also activate the AMP-activated protein kinase (AMPK), inhibiting the mammalian target of rapamycin (mTOR) signaling and curbing anabolic growth [47]. SCFAs enhance oxidative phosphorylation, shifting energy production from glycolysis and limiting tumor proliferation. In lipid metabolism, SCFAs suppress lipogenesis by downregulating the expression of sterol regulatory element-binding protein-1 (SREBP-1) and fatty acid synthase (FASN) [48,49], while promoting fatty acid oxidation (FAO) via peroxisome proliferator-activated receptor-gamma (PPAR-γ). These effects increase ATP production and reduce lipid accumulation, which is a characteristic feature in several aggressive, chemoresistant tumors [50]. SCFAs also prevent lipid droplet buildup, a hallmark of invasive cancers. SCFAs support mitochondrial function by fueling the tricarboxylic acid (TCA) cycle, boosting nicotinamide dehydrogenase “nicotinamide adenine dinucleotide (NAD) + hydrogen” (NADH) and ATP production, and restoring mitochondrial health in normal cells [51,52]. They regulate oxidative stress by increasing antioxidant enzymes superoxide dismutase (SOD) and glutathione peroxidase (GPx), protecting normal cells while enhancing ROS in tumor cells [53]. Additionally, SCFAs upregulate the expression of peroxisome proliferator-activated receptor γ coactivator 1 alpha (PGC-1α), promoting mitochondrial biogenesis and counteracting metabolic dysregulation [54]. 2.5. SCFAs and the Expression of miRNAs and lncRNAs in TME Beyond immune regulation, SCFAs, mainly butyrate, propionate, and acetate, are involved in regulating the expression of several gene sets in different cell types within the TME through HDAC inhibition and chromatin remodeling [55]. Among the genes that are regulated by SCFAs are several miRNAs and lncRNAs, which subsequently regulate tumor progression, immune evasion, and metabolic reprogramming [56,57]. For example, SCFAs have been reported to upregulate the expression of tumor-suppressive miRNAs like miR-34a, which inhibits Wnt/β-catenin and Notch signaling [58], miR-200 family, which blocks epithelial-mesenchymal transition (EMT) and enhances chemosensitivity [59], miR-146a, which reduces inflammation and M2 polarization [60], miR-124, which boosts CD8⁺ T cell cytotoxicity, and miR-155, which promotes M1 polarization and T cell activation [61]. Moreover, several SCFAs were reported to downregulate the expression of oncogenic miRNAs, including miR-21, miR-181a, miR-222/221, and miR-27a, all of which promote tumor growth, immune suppression, and/or metabolic reprogramming [62,63,64,65]. SCFAs also regulate lncRNAs by upregulating tumor suppressors like the maternally-expressed 3 gene (MEG3; induces apoptosis) [66], nuclear paraspeckle assembly transcript 1 (NEAT1; modulates immunity), growth arrest-specific transcript 5 (GAS5; enhances T cell function) [67], tumor suppressor candidate 7 (TUSC7; inhibits proliferation) [68], and long intergenic non-protein coding RNA, and p53 induced transcript (LINC-PINT; activates p53 signaling) [69]. At the same time, SCFAs have been reported to downregulate the expression of several oncogenic lncRNAs, including metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), HOX transcript antisense RNA (HOTAIR), small nucleolar RNA host gene 16 (SNHG16), urothelial cancer associated 1 (UCA1), and plasmacytoma variant translocation 1 (PVT1), which drive metastasis, immune evasion, and chemoresistance [70,71,72,73,74]. In short, SCFAs regulate key lncRNAs in the TME, contributing to tumor suppression by enhancing immunity and disrupting oncogenic signaling.
There is ample evidence to suggest that integrating SCFAs with immunotherapy enhances treatment outcomes in colorectal cancer (CRC), breast cancer, melanoma, lung cancer, and hepatocellular carcinoma (HCC), but not in glioblastoma, prostate, and ovarian cancer. This suggests that the anti-tumor effects precipitated by SCFAs could be cancer type-dependent, which raises the question of how cancers that respond positively to SCFA plus immunotherapy differ from those that do not. Focusing on this question may help identify cellular and molecular correlates that will inform future work on the therapeutic utility of SCFAs. One of the critical aspects to investigate here is the expression patterns of SCFA receptors and the integrity of the pathways initiated by SCFA receptor signaling in different types of cancer. Similarly related to this issue is whether specific effects precipitated by SCFAs in target cells of the TME can be assigned to specific SCFA receptors. SCFA-induced HDAC inhibition affects many genes, some of which play important roles in modulating the TME in favor of tumor regression. Searching for other genes that are subject to HDAC inhibition-dependent regulation should help in further appreciating the full extent of the anti-tumor effects of SCFAs within the TME. The absence of specific biomarkers for assessing the therapeutic value of SCFAs further complicates the issue. SCFAs play a crucial role in the differentiation and function of various immune cell subsets. The exact mechanism(s) underlying the immunomodulatory effects of SCFAs, particularly as they relate to macrophage polarization and Th cell differentiation, have yet to be elucidated. A detailed understanding of how SCFAs drive the differentiation of MDSCs is also pivotal to realistically assessing the risks of SCFA-based therapy in cancer patients. Exploring the therapeutic utility of SCFA-induced differentiation of MDSCs in inflammatory and autoimmune diseases also merits further investigation.
The gut microbiota metabolizes dietary fibers to produce SCFAs. Consumption of fiber-rich foods should, by definition, increase the concentration of SCFAs in the blood and tissues. However, determining the amount of fiber-rich food required to produce therapeutic levels of SCFAs remains unclear, making it difficult to calculate the appropriate dietary intake. Further, a detailed understanding of the pharmacokinetics and pharmacodynamics of SCFAs is still lacking. Moreover, tissue infiltration by SCFAs needs to be further investigated, especially in the case of hypoxic cores of solid tumors. Would too much SCFAs in the circulation and/or the TME overwhelm the anti-tumor immune response? Could insufficient SCFA levels contribute to immune tolerance and hinder rejection? Assessing the impact of dysbiosis resulting from antibiotics or chemotherapy, for example, on the type and concentration of SCFAs produced in cancer patients warrants further investigation.
| AMPK | AMP-Activated Protein Kinase |
| ARG1 | Arginase 1 |
| ATP | Adenosine Triphosphate |
| Bregs | Regulatory B Cells |
| CD | Cluster of Differentiation |
| CTL | Cytotoxic T Lymphocyte |
| CXCL1 | C-X-C Motif Chemokine Ligand 1 |
| DAMPs | Damage-Associated Molecular Patterns |
| DC | Dendritic Cell |
| EMT | Epithelial Mesenchymal Transition |
| FAO | Fatty Acid Oxidation |
| FASN | Fatty Acid Synthase |
| FFARs | Free Fatty Acid Receptors |
| Foxp3 | Forkhead Box P3 |
| GAS5 | Growth Arrest-Specific Transcript 5 |
| GPCRs | G-Protein coupled receptors |
| GPx | Glutathione Peroxidase |
| HDACs | Histone Deacetylases |
| HK2 | Hexokinase 2 (HK2) |
| HOTAIR | HOX transcript antisense RNA |
| IFN-γ | Interferon Gamma |
| IL | Interleukin |
| iNOS | Inducible Nitric Oxide Synthase |
| LAG-3 | Lymphocyte Activation Gene 3 |
| LINC-PINT | Long Intergenic Non-Protein Coding RNA, and p53 Induced Transcript |
| lncRNAs | Long Noncoding RNAs |
| M1 | Pro-Inflammatory |
| M2 | Anti-Inflammatory |
| MALAT1 | Metastasis-Associated Lung Adenocarcinoma Transcript 1 |
| MDSCs | Myeloid-Derived Suppressor Cells |
| MEG3 | Maternally-Expressed 3 Gene |
| MHC-II | Major Histocompatibility Complex-II |
| miRNAs | Micro RNAs |
| mTOR | Mammalian Target of Rapamycin |
| NADH | Nicotinamide Adenine Dinucleotide Hydrogen |
| NEAT1 | Nuclear Paraspeckle Assembly Transcript 1 |
| NET | Neutrophil Extracellular Trap |
| NK | Natural Killer |
| NKG2D | Natural Killer Group 2 Member D |
| NO | Nitric Oxide |
| PD-1 | Programmed Cell Death Protein 1 |
| PGC-1α | Peroxisome Proliferator-Activated Receptor γ Coactivator 1 Alpha |
| PKM2 | Pyruvate Kinase M2 |
| PPAR-γ | Peroxisome Proliferator-Activated Receptor-Gamma |
| PVT1 | Plasmacytoma Variant Translocation 1 |
| ROS | Reactive Oxygen Species |
| SCFAs | Short-Chain Fatty Acids |
| SNHG16 | Small Nucleolar RNA Host Gene 16 |
| SOD | Superoxide Dismutase |
| SREBP-1 | Sterol Regulatory Element-Binding Protein-1 |
| TAMPs | Tumor-Associated Molecular Patterns |
| TCA | Tricarboxylic Acid |
| TCR | T Cell Antigen Receptor |
| TGF-β | Tumor Growth Factor-Beta |
| Th | T Helper |
| TIM-3 | T-Cell Immunoglobulin and Mucin-Domain Containing-3 |
| TLRs | Toll-Like Receptors |
| TME | Tumor Microenvironment |
| TNF-α | Tumor Necrosis Factor-Alpha |
| Treg | Regulatory T Cell |
| TUSC7 | Tumor Suppressor Candidate 7 |
| UCA1 | Urothelial Cancer Associated 1 |
M.H. conceived the idea, B.R., M.W.M., A.S.J., T.M.K., and J.S.A.S. collected articles, prepared the first draft, B.R. and M.H. reviewed and edited the manuscript. All authors confirm that they have agreed to the submission and publication of this manuscript in [Cancer immunology connect].
The data used in this study were obtained from publicly available repositories: [PubMed, google scholar, google search]. No new datasets were generated during the current study.
The authors declare no conflicts of interest.
This research was funded by a competitive research grant (23010901133/MH), University of Sharjah, UAE.
The authors wish to acknowledge the generous in-house support of the Research Institute for Medical and Health Sciences, the University of Sharjah, UAE.
[1] De Visser, K.E.; Joyce, J.A. The Evolving Tumor Microenvironment: From Cancer Initiation to Metastatic Outgrowth. Cancer Cell 2023, 41, 374–403. [CrossRef] [PubMed]
[2] Li, C.; Yu, X.; Han, X.; Lian, C.; Wang, Z.; Shao, S.; Shao, F.; Wang, H.; Ma, S.; Liu, J. Innate Immune Cells in Tumor Microenvironment: A New Frontier in Cancer Immunotherapy. iScience 2024, 27, 110750. [CrossRef] [PubMed]
[3] Zhang, H.; Li, S.; Wang, D.; Liu, S.; Xiao, T.; Gu, W.; Yang, H.; Wang, H.; Yang, M.; Chen, P. Metabolic Reprogramming and Immune Evasion: The Interplay in the Tumor Microenvironment. Biomark. Res. 2024, 12. [CrossRef]
[4] Qu, S.; Gao, Y.; Ma, J.; Yan, Q. Microbiota-Derived Short-Chain Fatty Acids Functions in the Biology of B Lymphocytes: From Differentiation to Antibody Formation. Biomed. Pharmacother. 2023, 168. [CrossRef] [PubMed]
[5] Zhang, D.; Jian, Y.-P.; Zhang, Y.-N.; Li, Y.; Gu, L.-T.; Sun, H.-H.; Liu, M.-D.; Zhou, H.-L.; Wang, Y.-S.; Xu, Z.-X. Short-Chain Fatty Acids in Diseases. Cell Commun. Signal. 2023, 21, 212. [CrossRef]
[6] Zheng, M.; Zhang, W.; Chen, X.; Guo, H.; Wu, H.; Xu, Y.; He, Q.; Ding, L.; Yang, B. The Impact of Lipids on the Cancer–Immunity Cycle and Strategies for Modulating Lipid Metabolism to Improve Cancer Immunotherapy. Acta Pharm. Sin. B 2023, 13, 1488–1497. [CrossRef]
[7] Liu, H.; Wang, S.; Wang, J.; Guo, X.; Song, Y.; Fu, K.; Gao, Z.; Liu, D.; He, W.; Yang, L.-L. Energy Metabolism in Health and Diseases. Sig. Transduct. Target. Ther. 2025, 10, 1–71. [CrossRef]
[8] He, S.; Zheng, L.; Qi, C. Myeloid-Derived Suppressor Cells (MDSCs) in the Tumor Microenvironment and Their Targeting in Cancer Therapy. Mol. Cancer 2025, 24, 5. [CrossRef]
[9] Li, S.; Duan, Y.; Luo, S.; Zhou, F.; Wu, Q.; Lu, Z. Short-Chain Fatty Acids and Cancer. Trends Cancer 2025, 11, 154–168. [CrossRef]
[10] Yi, M.; Li, T.; Niu, M.; Mei, Q.; Zhao, B.; Chu, Q.; Dai, Z.; Wu, K. Exploiting Innate Immunity for Cancer Immunotherapy. Mol. Cancer 2023, 22, 187. [CrossRef]
[11] Huang, C.; Du, W.; Ni, Y.; Lan, G.; Shi, G. The Effect of Short-Chain Fatty Acids on M2 Macrophages Polarization in Vitro and in Vivo. Clin. Exp. Immunol. 2021, 207, 53–64. [CrossRef] [PubMed]
[12] Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and Tumor Progression: Signaling Pathways and Targeted Intervention. Signal. Transduct. Target Ther. 2021, 6, 263. [CrossRef] [PubMed]
[13] Zhou, D.; Li, Y. Gut Microbiota and Tumor-Associated Macrophages: Potential in Tumor Diagnosis and Treatment. Gut Microbes 2023, 15, 2276314. [CrossRef]
[14] Møller, S.H.; Wang, L.; Ho, P.-C. Metabolic Programming in Dendritic Cells Tailors Immune Responses and Homeostasis. Cell Mol. Immunol. 2022, 19, 370–383. [CrossRef]
[15] Nastasi, C.; Fredholm, S.; Willerslev-Olsen, A.; Hansen, M.; Bonefeld, C.M.; Geisler, C.; Andersen, M.H.; Ødum, N.; Woetmann, A. Butyrate and Propionate Inhibit Antigen-Specific CD8+ T Cell Activation by Suppressing IL-12 Production by Antigen-Presenting Cells. Sci. Rep. 2017, 7. [CrossRef]
[16] Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short Chain Fatty Acids Induce Both Effector and Regulatory T Cells by Suppression of Histone Deacetylases and Regulation of the mTOR-S6K Pathway. Mucosal Immunol. 2015, 8, 80–93. [CrossRef]
[17] Siemaszko, J.; Marzec-Przyszlak, A.; Bogunia-Kubik, K. NKG2D Natural Killer Cell Receptor—A Short Description and Potential Clinical Applications. Cells 2021, 10. [CrossRef] [PubMed]
[18] Kim, C.H. Complex Regulatory Effects of Gut Microbial Short-Chain Fatty Acids on Immune Tolerance and Autoimmunity. Cell. Mol. Immunol. 2023, 20, 341–350. [CrossRef]
[19] Altamura, S.; Lombardi, F.; Palumbo, P.; Cinque, B.; Ferri, C.; Del Pinto, R.; Pietropaoli, D. The Evolving Role of Neutrophils and Neutrophil Extracellular Traps (NETs) in Obesity and Related Diseases: Recent Insights and Advances. Int. J. Mol. Sci. 2024, 25. [CrossRef]
[20] Yan, Q.; Jia, S.; Li, D.; Yang, J. The Role and Mechanism of Action of Microbiota-Derived Short-Chain Fatty Acids in Neutrophils: From the Activation to Becoming Potential Biomarkers. Biomed. Pharmacother. 2023, 169. [CrossRef]
[21] Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10. [CrossRef]
[22] Liu, X.; Shao, J.; Liao, Y.-T.; Wang, L.-N.; Jia, Y.; Dong, P.; Liu, Z.; He, D.; Li, C.; Zhang, X. Regulation of Short-Chain Fatty Acids in the Immune System. Front. Immunol. 2023, 14. [CrossRef]
[23] Abdelhalim, K.A. Short-Chain Fatty Acids (SCFAs) from Gastrointestinal Disorders, Metabolism, Epigenetics, Central Nervous System to Cancer—A Mini-Review. Chem.-Biol. Interact. 2024, 388. [CrossRef] [PubMed]
[24] Yu, X.; Li, W.; Li, Z.; Wu, Q.; Sun, S. Influence of Microbiota on Tumor Immunotherapy. Int. J. Biol. Sci. 2024, 20, 2264–2294. [CrossRef]
[25] Kim, C.H.; Park, J.; Kim, M. Gut Microbiota-Derived Short-Chain Fatty Acids, T Cells, and Inflammation. Immune Netw. 2014, 14, 277–288. [CrossRef]
[26] Ney, L.-M.; Wipplinger, M.; Grossmann, M.; Engert, N.; Wegner, V.D.; Mosig, A.S. Short Chain Fatty Acids: Key Regulators of the Local and Systemic Immune Response in Inflammatory Diseases and Infections. Open Biol. 2023, 13. [CrossRef]
[27] Qiu, Y.; Ke, S.; Chen, J.; Qin, Z.; Zhang, W.; Yuan, Y.; Meng, D.; Zhao, G.; Wu, K.; Li, B.; et al. FOXP3+ Regulatory T Cells and the Immune Escape in Solid Tumours. Front. Immunol. 2022, 13. [CrossRef]
[28] Rumiano, L.; Manzo, T. Lipids Guide T Cell Antitumor Immunity by Shaping Their Metabolic and Functional Fitness. Trends Endocrinol. Metab. 2024, . [CrossRef]
[29] Wang, Y.; Wang, Y.; Ren, Y.; Zhang, Q.; Yi, P.; Cheng, C. Metabolic Modulation of Immune Checkpoints and Novel Therapeutic Strategies in Cancer. Semin. Cancer Biol. 2022, 86, 542–565. [CrossRef]
[30] Overacre-Delgoffe, A.E.; Hand, T.W. Regulation of Tissue-Resident Memory T Cells by the Microbiota. Mucosal Immunol. 2022, 15, 408–417. [CrossRef]
[31] Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [CrossRef] [PubMed]
[32] Zhang, W.; Mackay, C.R.; Gershwin, M.E. Immunomodulatory Effects of Microbiota-Derived Short-Chain Fatty Acids in Autoimmune Liver Diseases. J. Immunol. 2023, 210, 1629–1639. [CrossRef] [PubMed]
[33] Yang, Y.; Li, C.; Liu, T.; Dai, X.; Bazhin, A.V. Myeloid-Derived Suppressor Cells in Tumors: From Mechanisms to Antigen Specificity and Microenvironmental Regulation. Front. Immunol. 2020, 11. [CrossRef]
[34] Wang, H.; Zhou, F.; Qin, W.; Yang, Y.; Li, X.; Liu, R. Metabolic Regulation of Myeloid-Derived Suppressor Cells in Tumor Immune Microenvironment: Targets and Therapeutic Strategies. Theranostics 2025, 15, 2159–2184. [CrossRef]
[35] Radojević, D.; Bekić, M.; Gruden-Movsesijan, A.; Ilić, N.; Dinić, M.; Bisenić, A.; Golić, N.; Vučević, D.; Đokić, J.; Tomić, S. Myeloid-Derived Suppressor Cells Prevent Disruption of the Gut Barrier, Preserve Microbiota Composition, and Potentiate Immunoregulatory Pathways in a Rat Model of Experimental Autoimmune Encephalomyelitis. Gut Microbes 2022, 14, 2127455. [CrossRef]
[36] Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of Short-Chain Fatty Acids and Their Receptors in Inflammation and Carcinogenesis. Pharmacol. Ther. 2016, 164, 144–151. [CrossRef]
[37] Fatty Acid Metabolism of Immune Cells: A New Target of Tumour Immunotherapy | Cell Death Discovery. Available online: https://www.nature.com/articles/s41420-024-01807-9 (accessed on 26 March 2025).
[38] Fultang, N.; Li, X.; Li, T.; Chen, Y.H. Myeloid-Derived Suppressor Cell Differentiation in Cancer: Transcriptional Regulators and Enhanceosome-Mediated Mechanisms. Front. Immunol. 2021, 11. [CrossRef]
[39] Yan, D.; Adeshakin, A.O.; Xu, M.; Afolabi, L.O.; Zhang, G.; Chen, Y.H.; Wan, X. Lipid Metabolic Pathways Confer the Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor. Front. Immunol. 2019, 10. [CrossRef]
[40] Wang, R.; Li, B.; Huang, B.; Li, Y.; Liu, Q.; Lyu, Z.; Chen, R.; Qian, Q.; Liang, X.; Pu, X.; et al. Gut Microbiota-Derived Butyrate Induces Epigenetic and Metabolic Reprogramming in Myeloid-Derived Suppressor Cells to Alleviate Primary Biliary Cholangitis. Gastroenterology 2024, 167, 733–749.e3. [CrossRef]
[41] Guerra, L.; Bonetti, L.; Brenner, D. Metabolic Modulation of Immunity: A New Concept in Cancer Immunotherapy. Cell Rep. 2020, 32, 107848. [CrossRef] [PubMed]
[42] Optimizing CD8+ T Cell-Based Immunotherapy via Metabolic Interventions: A Comprehensive Review of Intrinsic and Extrinsic Modulators | Experimental Hematology & Oncology | Full Text. Available online: https://ehoonline.biomedcentral.com/articles/10.1186/s40164-024-00575-7 (accessed on 26 March 2025).
[43] Tufail, M.; Jiang, C.-H.; Li, N. Altered Metabolism in Cancer: Insights into Energy Pathways and Therapeutic Targets. Mol. Cancer 2024, 23, 203. [CrossRef]
[44] Gomes, S.; Rodrigues, A.C.; Pazienza, V.; Preto, A. Modulation of the Tumor Microenvironment by Microbiota-Derived Short-Chain Fatty Acids: Impact in Colorectal Cancer Therapy. Int. J. Mol. Sci. 2023, 24. [CrossRef] [PubMed]
[45] Campos-Perez, W.; Martinez-Lopez, E. Effects of Short Chain Fatty Acids on Metabolic and Inflammatory Processes in Human Health. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866. [CrossRef]
[46] Wang, G.; Yu, Y.; Wang, Y.; Wang, J.; Guan, R.; Sun, Y.; Shi, F.; Gao, J.; Fu, X. Role of SCFAs in Gut Microbiome and Glycolysis for Colorectal Cancer Therapy. J. Cell. Physiol. 2019, 234, 17023–17049. [CrossRef]
[47] An, H.; Jang, Y.; Choi, J.; Hur, J.; Kim, S.; Kwon, Y. New Insights into AMPK, as a Potential Therapeutic Target in Metabolic Dysfunction-Associated Steatotic Liver Disease and Hepatic Fibrosis. Biomol. Ther. 2025, 33, 18–38. [CrossRef] [PubMed]
[48] Huang, K.; Han, Y.; Chen, Y.; Shen, H.; Zeng, S.; Cai, C. Tumor Metabolic Regulators: Key Drivers of Metabolic Reprogramming and the Promising Targets in Cancer Therapy. Mol. Cancer 2025, 24, 7. [CrossRef]
[49] Feng, T.; Zhang, H.; Zhou, Y.; Zhu, Y.; Shi, S.; Li, K.; Lin, P.; Chen, J. Roles of Posttranslational Modifications in Lipid Metabolism and Cancer Progression. Biomark. Res. 2024, 12. [CrossRef]
[50] Chowdhury, P.S.; Chamoto, K.; Kumar, A.; Honjo, T. PPAR-Induced Fatty Acid Oxidation in T Cells Increases the Number of Tumor-Reactive CD8+ T Cells and Facilitates Anti-PD-1 Therapy. Cancer Immunol. Res. 2018, 6, 1375–1387. [CrossRef] [PubMed]
[51] González-Bosch, C.; Zunszain, P.A.; Mann, G.E. Control of Redox Homeostasis by Short-Chain Fatty Acids: Implications for the Prevention and Treatment of Breast Cancer. Pathogens 2023, 12. [CrossRef]
[52] Schönfeld, P.; Wojtczak, L. Short- and Medium-Chain Fatty Acids in Energy Metabolism: The Cellular Perspective. J. Lipid Res. 2016, 57, 943–954. [CrossRef]
[53] Ferrer, M.; Buey, B.; Grasa, L.; Mesonero, J.E.; Latorre, E. Protective Role of Short-Chain Fatty Acids on Intestinal Oxidative Stress Induced by TNF-α. Cell Stress Chaperones 2024, 29, 769–776. [CrossRef] [PubMed]
[54] Ghoneum, A.; Abdulfattah, A.Y.; Warren, B.O.; Shu, J.; Said, N. Redox Homeostasis and Metabolism in Cancer: A Complex Mechanism and Potential Targeted Therapeutics. Int. J. Mol. Sci. 2020, 21. [CrossRef] [PubMed]
[55] Licciardi, P.V.; Wong, S.-S.; Tang, M.L.; Karagiannis, T.C. Epigenome Targeting by Probiotic Metabolites. Gut Pathog. 2010, 2, 24. [CrossRef] [PubMed]
[56] Zhang, Y.; Mao, Q.; Xia, Q.; Cheng, J.; Huang, Z.; Li, Y.; Chen, P.; Yang, J.; Fan, X.; Liang, Y.; et al. Noncoding RNAs Link Metabolic Reprogramming to Immune Microenvironment in Cancers. J. Hematol. Oncol. 2021, 14, 169. [CrossRef]
[57] Liu, X.; Zhao, S.; Sui, H.; Liu, H.; Yao, M.; Su, Y.; Qu, P. MicroRNAs/LncRNAs Modulate MDSCs in Tumor Microenvironment. Front. Oncol. 2022, 12. [CrossRef]
[58] Li, Q.; Geng, S.; Luo, H.; Wang, W.; Mo, Y.-Q.; Luo, Q.; Wang, L.; Song, G.-B.; Sheng, J.-P.; Xu, B. Signaling Pathways Involved in Colorectal Cancer: Pathogenesis and Targeted Therapy. Sig. Transduct. Target. Ther. 2024, 9, 1–48. [CrossRef]
[59] Supic, G.; Wagner, D.; Magic, Z. Epigenetic Impact of Bioactive Dietary Compounds in Cancer Chemoprevention. In Critical Dietary Factors in Cancer Chemoprevention , Ullah, M.F.; Ahmad, A., Eds.; Springer International Publishing: Cham, Switzerland, 2016; 153–181. ISBN 978-3-319-21461-0. . [CrossRef]
[60] Padoan, A.; Plebani, M.; Basso, D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int. J. Mol. Sci. 2019, 20. [CrossRef]
[61] Giammona, A.; Galuzzi, B.G.; Imperia, E.; Gervasoni, C.; Remedia, S.; Restaneo, L.; Nespoli, M.; De Gara, L.; Tani, F.; Cicala, M.; et al. Chronic Gastrointestinal Disorders and miRNA-Associated Disease: An Up-to-Date. Int. J. Mol. Sci. 2025, 26. [CrossRef]
[62] Guz, M.; Jeleniewicz, W.; Malm, A.; Korona-Glowniak, I. A Crosstalk between Diet, Microbiome and microRNA in Epigenetic Regulation of Colorectal Cancer. Nutrients 2021, 13. [CrossRef]
[63] Bi, K.; Zhang, X.; Chen, W.; Diao, H. MicroRNAs Regulate Intestinal Immunity and Gut Microbiota for Gastrointestinal Health: A Comprehensive Review. Genes 2020, 11. [CrossRef]
[64] Parasramka, M.A.; Ho, E.; Williams, D.E.; Dashwood, R.H. MicroRNAs, Diet, and Cancer: New Mechanistic Insights on the Epigenetic Actions of Phytochemicals. Mol. Carcinog. 2012, 51, 213–230. [CrossRef] [PubMed]
[65] Li, W.; Lu, Y.; Ye, C.; Ouyang, M. The Regulatory Network of MicroRNA in the Metabolism of Colorectal Cancer. J. Cancer 2021, 12, 7454–7464. [CrossRef] [PubMed]
[66] Olmedo-Suárez, M.Á.; Ramírez-Díaz, I.; Pérez-González, A.; Molina-Herrera, A.; Coral-García, M.Á.; Lobato, S.; Sarvari, P.; Barreto, G.; Rubio, K. Epigenetic Regulation in Exposome-Induced Tumorigenesis: Emerging Roles of ncRNAs. Biomolecules 2022, 12. [CrossRef]
[67] Yuan, H.; Xu, R.; Li, S.; Zheng, M.; Tong, Q.; Xiang, M.; Zhang, Y. The Malignant Transformation of Viral Hepatitis to Hepatocellular Carcinoma: Mechanisms and Interventions. MedComm 2025, 6, e70121. [CrossRef]
[68] Xu, J.; Zhang, R.; Zhao, J. The Novel Long Noncoding RNA TUSC7 Inhibits Proliferation by Sponging MiR-211 in Colorectal Cancer. Cell. Physiol. Biochem. 2017, 41, 635–644. [CrossRef]
[69] Ungkulpasvich, U.; Hatakeyama, H.; Hirotsu, T.; di Luccio, E. Pancreatic Cancer and Detection Methods. Biomedicines 2023, 11. [CrossRef]
[70] Mohammadpour, S.; Torshizi Esfahani, A.; Sarpash, S.; Vakili, F.; Zafarjafarzadeh, N.; Mashaollahi, A.; Pardakhtchi, A.; Nazemalhosseini-Mojarad, E. Hippo Signaling Pathway in Colorectal Cancer: Modulation by Various Signals and Therapeutic Potential. Anal. Cell. Pathol. 2024, 2024, 5767535. [CrossRef]
[71] Fan, S.; Xing, J.; Jiang, Z.; Zhang, Z.; Zhang, H.; Wang, D.; Tang, D. Effects of Long Non-Coding RNAs Induced by the Gut Microbiome on Regulating the Development of Colorectal Cancer. Cancers 2022, 14. [CrossRef]
[72] Lu, Q.; Liang, Y.; Meng, X.; Zhao, Y.; Fan, H.; Hou, S. The Role of Long Noncoding RNAs in Intestinal Health and Diseases: A Focus on the Intestinal Barrier. Biomolecules 2023, 13. [CrossRef]
[73] Huo, M.; Zhang, J.; Huang, W.; Wang, Y. Interplay among Metabolism, Epigenetic Modifications, and Gene Expression in Cancer. Front. Cell Dev. Biol. 2021, 9. [CrossRef]
[74] Mecca, M.; Picerno, S.; Cortellino, S. The Killer’s Web: Interconnection between Inflammation, Epigenetics and Nutrition in Cancer. Int. J. Mol. Sci. 2024, 25. [CrossRef] [PubMed]
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more