
APA Style
Soumi Das. (2025). Unraveling Conformational Thermodynamics of Ligand Binding to Fluoride Riboswitch Aptamer: Implications for Therapeutic Design. Molecular Modeling Connect, 2 (Article ID: 0012). https://doi.org/Registering DOIMLA Style
Soumi Das. "Unraveling Conformational Thermodynamics of Ligand Binding to Fluoride Riboswitch Aptamer: Implications for Therapeutic Design". Molecular Modeling Connect, vol. 2, 2025, Article ID: 0012, https://doi.org/Registering DOI.Chicago Style
Soumi Das. 2025. "Unraveling Conformational Thermodynamics of Ligand Binding to Fluoride Riboswitch Aptamer: Implications for Therapeutic Design." Molecular Modeling Connect 2 (2025): 0012. https://doi.org/Registering DOI.
 ACCESS
Research Article
ACCESS
Research Article
Volume 2, Article ID: 2025.0012

Soumi Das
soumi.das@bose.res.in
Department of Physics of Complex Systems, S. N. Bose National Centre for Basic Sciences, Salt Lake, Kolkata 700106, India
Received: 13 Jun 2025 Accepted: 01 Oct 2025 Available Online: 08 Oct 2025
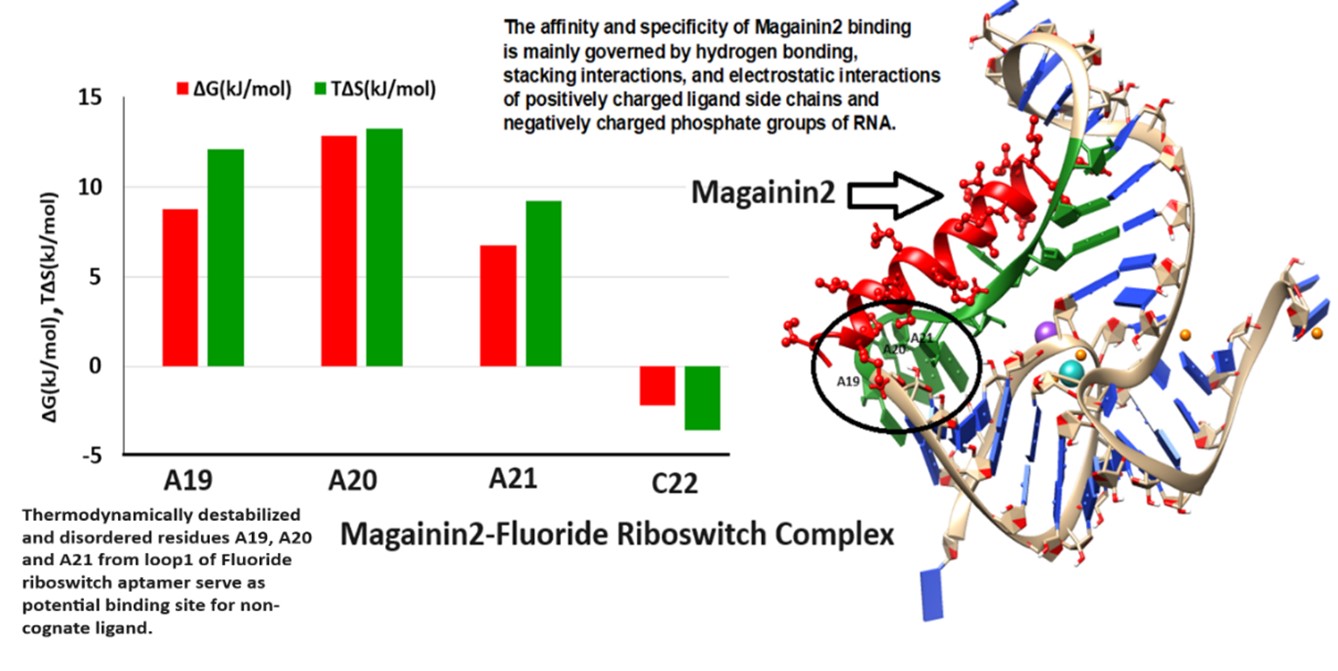
Riboswitches are structured non-coding mRNA segments in which ligand binding to an aptamer domain induces conformational changes in a downstream expression platform to regulate gene expression, positioning them as attractive targets for antimicrobial therapy. The fluoride riboswitch, found in several pathogenic bacteria, is a promising yet underexplored target despite evidence of its role in bacterial defense through fluoride ion (F⁻) binding in the presence of Mg²⁺. In this study, we investigate the conformational stability of (i) the holo form of the Thermotoga petrophila fluoride riboswitch aptamer (RNA in the presence of F⁻+Mg²⁺+K⁺) relative to (ii) the apo form (RNA in the absence of F⁻+Mg²⁺+K⁺). Conformational thermodynamic analysis reveals that the holo riboswitch is stabilized by the Ion recognition site, the Pseudoknot, and Stem 1, whereas Stem 2, Loop 1, Loop 2, and most unpaired nucleotides exhibit pronounced disorder and destabilization. Complementary docking studies identify these destabilized regions as putative binding pockets for non-cognate ligands. Together, these findings provide structural and thermodynamic insights into fluoride riboswitch–ligand interactions, guiding the design of nucleic acid–targeted therapeutics, including RNA-modulating drugs and engineered aptamers. Future in vitro and in vivo validation will be critical for translating these computational predictions into novel strategies to combat antimicrobial resistance.
Disclaimer: This is not the final version of the article. Changes may occur when the manuscript is published in its final format.
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more