
APA Style
Kholmirzo T. Kholmurodov, Ivan A. Baigunov, Pavel P. Gladyshev, Mirzoaziz A. Husenzoda, Hanan G. Elhaes, Medhat A. Ibrahim. (2025). Unusual Arrangement of Catalytic Loops of the Alcohol Dehydrogenase Enzyme During the Adsorption Process on a Graphitic Carbon Surface. Molecular Modeling Connect, 2 (Article ID: 0003). https://doi.org/10.69709/MolModC.2025.101003MLA Style
Kholmirzo T. Kholmurodov, Ivan A. Baigunov, Pavel P. Gladyshev, Mirzoaziz A. Husenzoda, Hanan G. Elhaes, Medhat A. Ibrahim. "Unusual Arrangement of Catalytic Loops of the Alcohol Dehydrogenase Enzyme During the Adsorption Process on a Graphitic Carbon Surface". Molecular Modeling Connect, vol. 2, 2025, Article ID: 0003, https://doi.org/10.69709/MolModC.2025.101003.Chicago Style
Kholmirzo T. Kholmurodov, Ivan A. Baigunov, Pavel P. Gladyshev, Mirzoaziz A. Husenzoda, Hanan G. Elhaes, Medhat A. Ibrahim. 2025. "Unusual Arrangement of Catalytic Loops of the Alcohol Dehydrogenase Enzyme During the Adsorption Process on a Graphitic Carbon Surface." Molecular Modeling Connect 2 (2025): 0003. https://doi.org/10.69709/MolModC.2025.101003.
 ACCESS
Research Article
ACCESS
Research Article
Volume 2, Article ID: 2025.0003

Kholmirzo T. Kholmurodov
kholmirzo@gmail.com

Ivan A. Baigunov

Pavel P. Gladyshev

Mirzoaziz A. Husenzoda

Hanan G. Elhaes

Medhat A. Ibrahim
1 Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research, 141980 Dubna, Moscow Region, Russia
2 Department of Chemistry, New Technologies and Materials, Dubna State University, 141980 Dubna, Moscow Region, Russia
3 Department of Fundamental Nuclear Interactions, Faculty of Physics, Lomonosov Moscow State University, 119991 Moscow, Russia
4 S.U. Umarov Physical-Technical Institute (PhTI), Aini Ave. 299/1, Dushanbe 734063, Tajikistan
5 Department of Technical Operation of Air Transport, Faculty of Transport and Road Infrastructure, Tajik Technical University, Academicians Radjabov str., 10, Dushanbe 734042, Tajikistan
6 Physics Department, Faculty of Women for Arts, Science and Education, Ain Shams University, Cairo 11757, Egypt
7 Spectroscopy Department, National Research Centre, 33 El-Bohouth St., Dokki, Giza 12622, Egypt
8 Molecular Modeling and Spectroscopy Laboratory, Centre of Excellence for Advanced Science, National Research Centre, 33 El-Bohouth St., Dokki, Giza 12622, Egypt
* Author to whom correspondence should be addressed
Received: 26 Sep 2024 Accepted: 14 Feb 2025 Available Online: 14 Feb 2025 Published: 20 Mar 2025
Molecular dynamics simulations (MDs) were conducted to investigate the alcohol dehydrogenase (ADH) enzyme and its cofactor, nicotinamide adenine dinucleotide (NAD), solvated in an aqueous system on a graphite-carbon surface. The study focuses on key structural aspects of ADH, particularly its conformational changes, including the rotation of the catalytic domain, the coenzyme binding domain, and the rearrangement of the active center required for catalytic activation. MDs were performed for 100 ns to track the conformational and rotational changes of the ADH+NAD complex in an aqueous environment. The diffusion process of ADH+NAD was observed for approximately 50–60 ns, after which the complex made contact with the graphite surface, leading to final relaxation. Upon adsorption of ADH onto the carbon surface, NAD exhibited fluctuations between 55 and 60 ns, coinciding with the opening of two catalytic loops associated with the NAD-binding region. Experimental and kinetic studies suggest that these structural changes may significantly impact the enzymatic activity of ADH in the presence of a carbon surface.
The enzyme alcohol dehydrogenase (ADH) of yeast is one of the interesting objects in the chemical, biological, pharmaceutical, electrochemical, agrochemical and aromatic industries [1,2,3,4,5]. For example, in [5], a successful preparation of hexanal catalyzed by ADH from baker’s yeast was carried out. Essential to the functionality of ADH is that it must be added in stoichiometric amounts and cannot be replaced by more economical synthetic products. At the same time, the coenzyme NAD+ (nicotinamide adenine dinucleotide) is widely used in biocatalytic oxidations catalyzed by ADH. The most intriguing in the structural aspect of ADH, as shown by X-ray crystallography, is that ADH undergoes global conformational changes (it does not matter when binding the NAD+—oxidized form or the NADH—reduced form of NAD), including rotation of the catalytic domain relative to the coenzyme binding domain and rearrangement of the active center to form a catalytically active enzyme. A change in conformation requires the presence of a complete coenzyme and depends on various chemical or mutational substitutions. These substitutions can enhance catalytic activity by altering the kinetics of isomerization and the rate of coenzyme dissociation [2,3,4,5,6]. As for the structural aspect of the enzyme, the orientation of ADH on various sorbents and electrically conductive matrices was studied using experimental observations and mathematical modeling of the protein dependence on the pH of the solution. As is known, the deactivation of the enzyme is caused by unsuitable conditions (temperature, pH), which leads to a change in the activity of the enzyme ADH. In relation to the problems of modeling the effect of pH on the orientation of protein sorption, the sorption behavior of the ADH enzyme based on specific interactions of protein groups with groups on the surface and its charge was previously studied in [7,8,9,10,11,12,13,14,15]. It was assumed that the conformation of the ADH enzyme corresponds to its conformation in the crystal structure and does not change when the surrounding solution and the surface of the sorbent change, which is a rather rough approximation. The implementation of various options includes, for example, the immobilization of a two-substrate enzyme on the surface of electrode materials [7,8,9,10,11,12,13,14,15]. However, it should be noted that the experimental study of the above issues is difficult. Therefore, in recent years, computational and simulation analysis methods have been widely used for these purposes [16,17,18,19,20,21,22,23]. This study utilized computer molecular dynamics (MD) modeling to study the structural conformational changes of the enzyme ADH and its cofactor NAD, which occur in aqueous solution when interacting with a graphite carbon surface. The MD analysis data provide a significant extension of the initial basic model, thereby allowing the protein conformation to be changed in the region of titrated amino acid residues of ADH in atomic/molecular details. The identification of the characteristic conformation of key titrated amino acids may become a necessary stage for further research and the implementation of a numerical experiment, which will be carried out by varying the pH and surface charge values [2,7,8,9,10,11,12,13,14,15]. Of course, the current research correlates with several parallel theoretical and experimental works concerning the immobilization and adsorption of enzymes on a carbon surface, the location and fixation of enzymes on carbon platforms, as well as on the surfaces of bioanodes, etc. It is worth noting that the choice of carbon surfaces and platforms is due to good control of the location of enzymes on the surface with very low enzyme consumption. To date, the kinetic behavior of the ADH enzyme in a solution fixed on carbon platforms has been studied. However, there are few comparative studies of the ADH enzyme that take into account the kinetic behavior of the enzyme, despite the studies of the structure and kinetic mechanism of immobilized enzymes available in the literature. Kinetic parameters obtained for the ADH enzyme attached to a carbon surface (carbon fiber paper) indicate a decrease in activity after the immobilization and fixation process. A significant loss of enzymatic activity was observed after immobilization, despite the retention of the enzyme’s affinity for its substrates and coenzymes. It is believed that MD observations of the structural behavior of ADH+NAD, diffusion and adsorption processes correlate well with the kinetic parameters obtained for the enzyme fixed with ADH on a carbon platform. Various kinetic parameters (mainly the concentration of substrates and coenzyme) indicate a change (loss or decrease) in enzymatic activity of the immobilized enzymes following the surface immobilization process. In particular, the observations obtained in the current study may correlate well with practical applications, for example, with the growing interest of biotechnologists in the use of immobilized enzymes such as ADH for many purposes, bioremediation, sensors and biofuel elements, etc.
2.1. Molecular Dynamics Simulations In this section, we have implemented the basic MD-AMBER production simulation (CPU/GPU) (also common with many other types of simulation based on processors and GPUs to perform MD simulation with AMBER). Molecular dynamics (MD) modeling has been used with the Amber 18 code (CPU/GPU environment) for the PDB ID: 3COS of the crystal structure of human alcohol dehydrogenase Class II [24]. A detailed description of the main parameters and algorithms used in computer modeling of molecular dynamics is given in the previously published article in [25]. For the ADH + NAD + water + graphite carbon surface system, the general molecular structure shown below in Figure 1a,b was obtained almost manually, step by step. The simulated system posed no particular difficulties in preparation stage, as it did not require special software or a force field database beyond the MD-AMBER computing environment. Thus, to keep the surface atom positions fixed we have used [25] the restraint options (ntr = 1, restraint_wt = 25, restraintmask = ‘:383–35140’), so far, the positional restraint was varied from 5 to 25 kcal mol–1 Å–2 for the several set of the MD calculations. It’s worth noting that the ntr=1 means that position restraints have been activated. =. Consequently, the atoms (graphitic carbon surfaces) to be restrained, along with the force constant, must be specified via the GROUP input. In the MD run, after specification of the namelists, a title is given, followed by the force constant for the restraint (in kcal mol–1 Å–2) and then a specification of surface carbon (C) residues or atoms to restrain. Residues can be specified using the “RES” keyword. We have chosen a force constant of 25 kcal mol–1 Å–2 and restrained the surrounding ADH+NAD + water confined C-surfaces residues were through 383–35140 (from 1 to 382 the correspondent numerations stand for the ADH+NAD enzyme). The total number of atoms for the ADH+NAD + water + carbon surfaces were 157,371. It’s worth also noting that the ADH is a Zn (II)-bound metalloenzyme, and during the adsorption process in ADH the position of the metal bound to Zn (II) was fixed, as in the original PDB file (ID: 3COS of the crystal structure of human alcohol dehydrogenase class II ADH). It was necessary to maintain the position of Zn (II) inside the enzyme pocket of ADH+NAD, thereby, restraining the relative position of the metal bound to Zn (II) during the entire adsorption process. So far, the position of the Zn (II)-bound metal has not been violated, since the main attention has been paid to the orientation sorption of the ADH+NAD and the behavior of the NAD co-enzyme, due to the conformational behavior of the catalytic loops of the enzyme relaxed equilibrated structure. The MD modeling on the molecular system of ADH+NAD+water+carbon surface, equilibrated at a target temperature of 303 K, was carried out in three stages [25]: energy minimization, NVT and NPT relaxation procedures (Figure 1).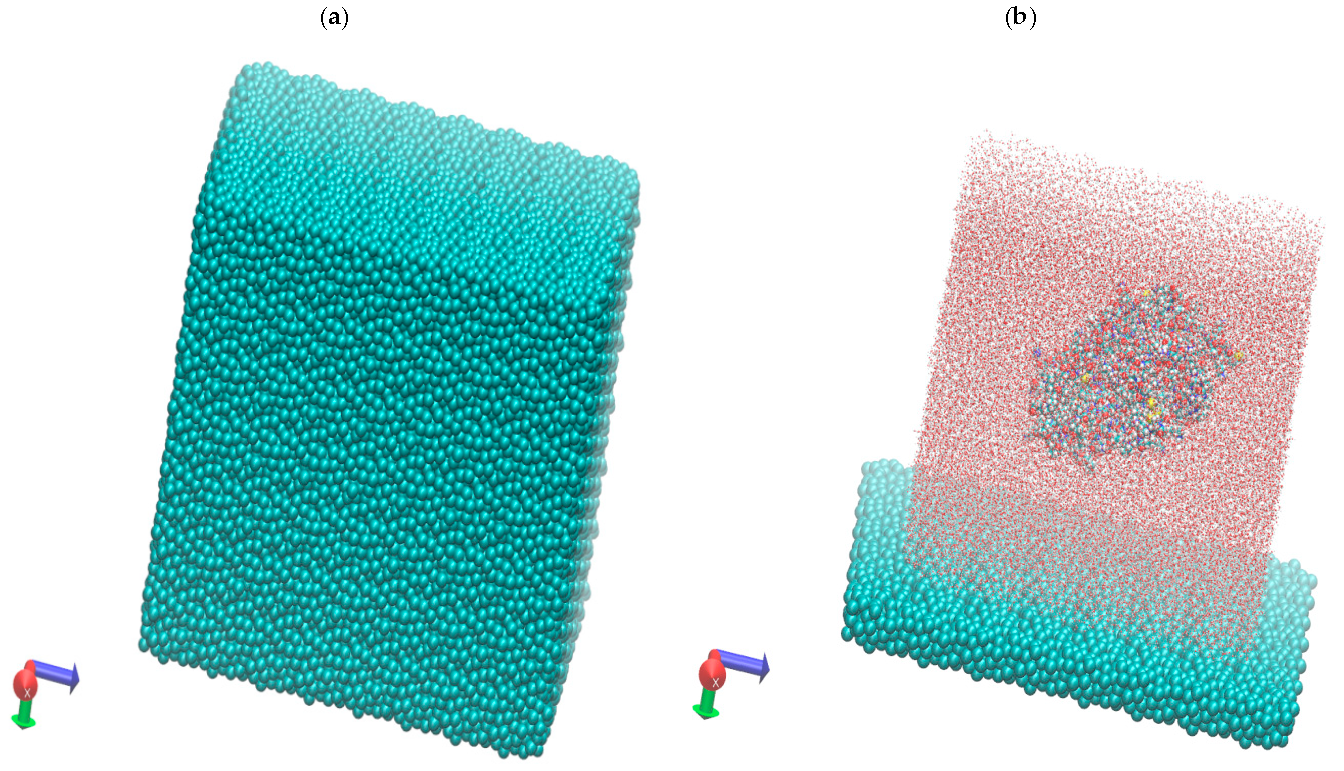
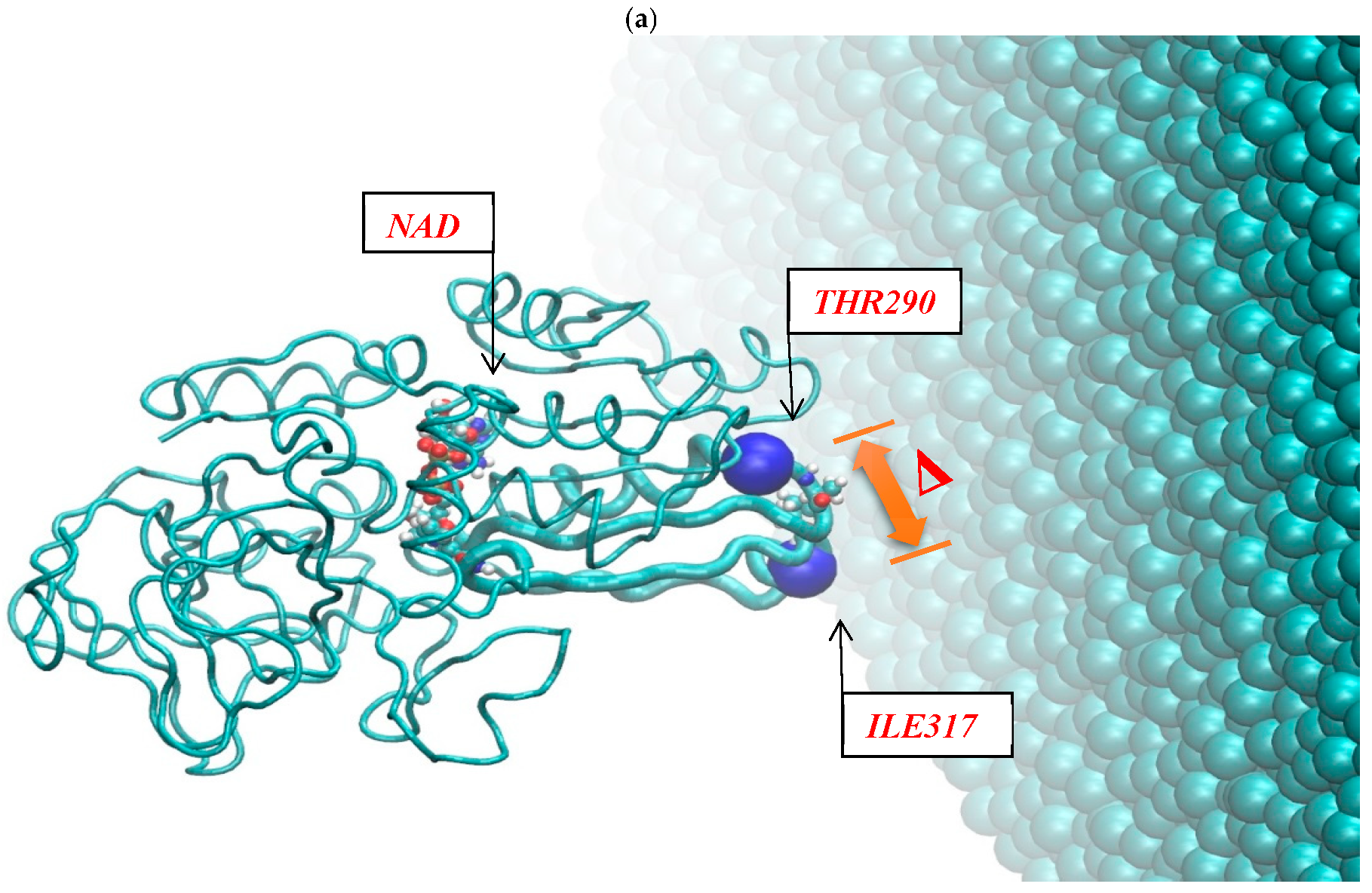
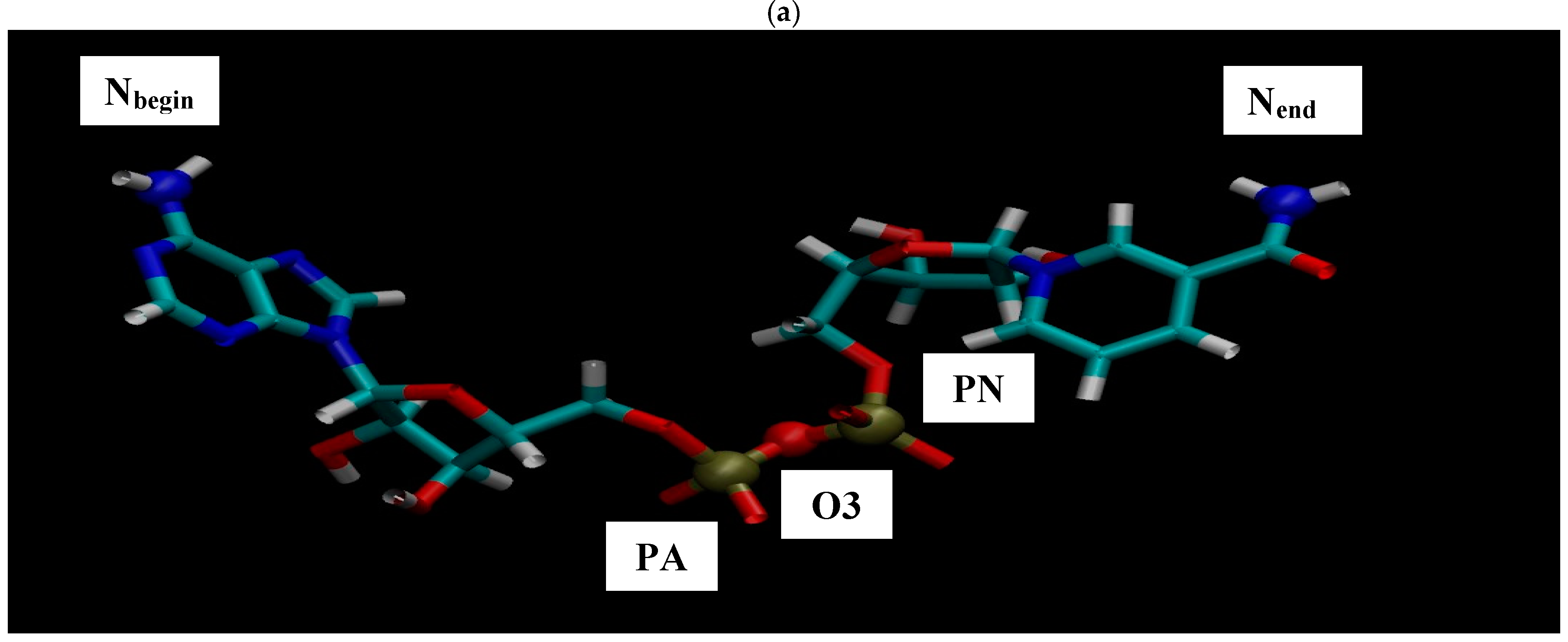
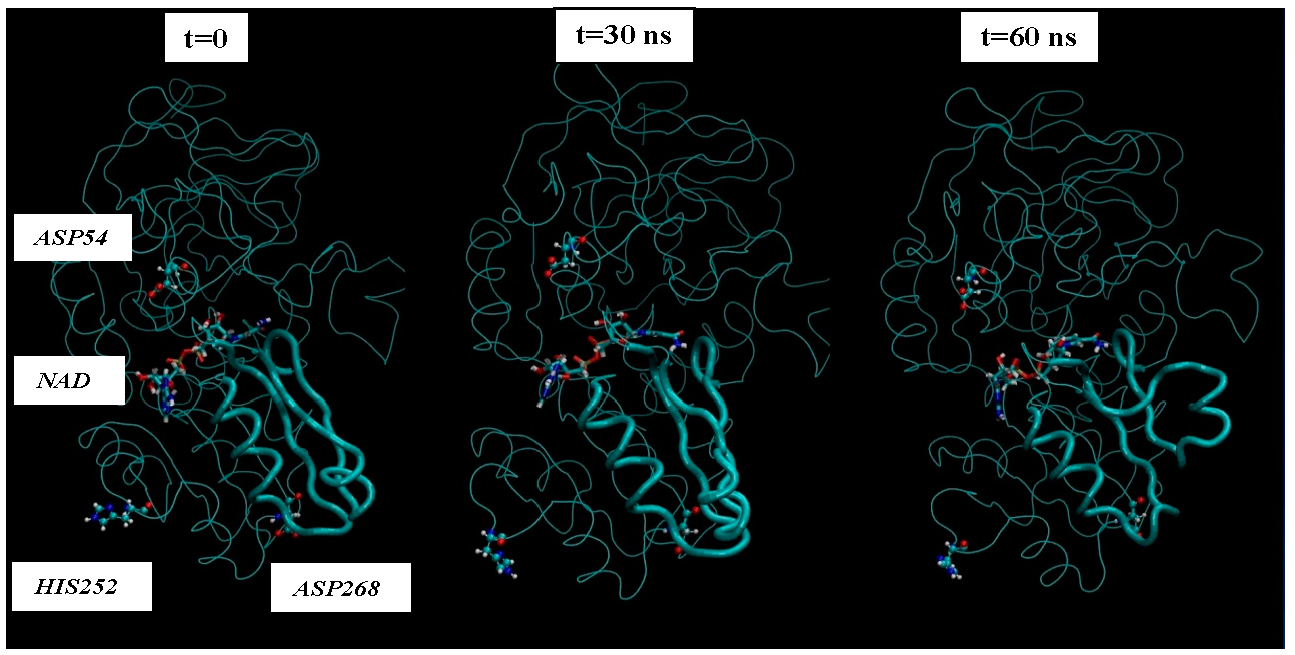
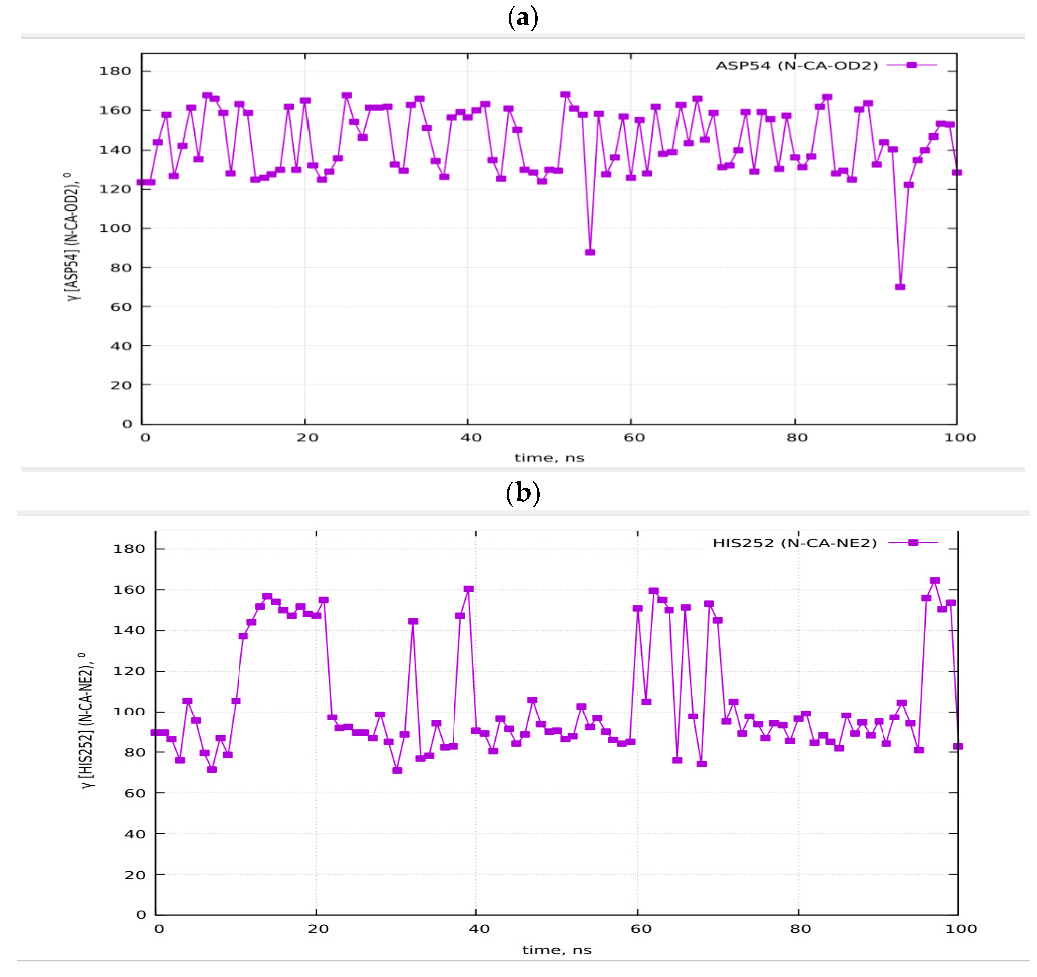
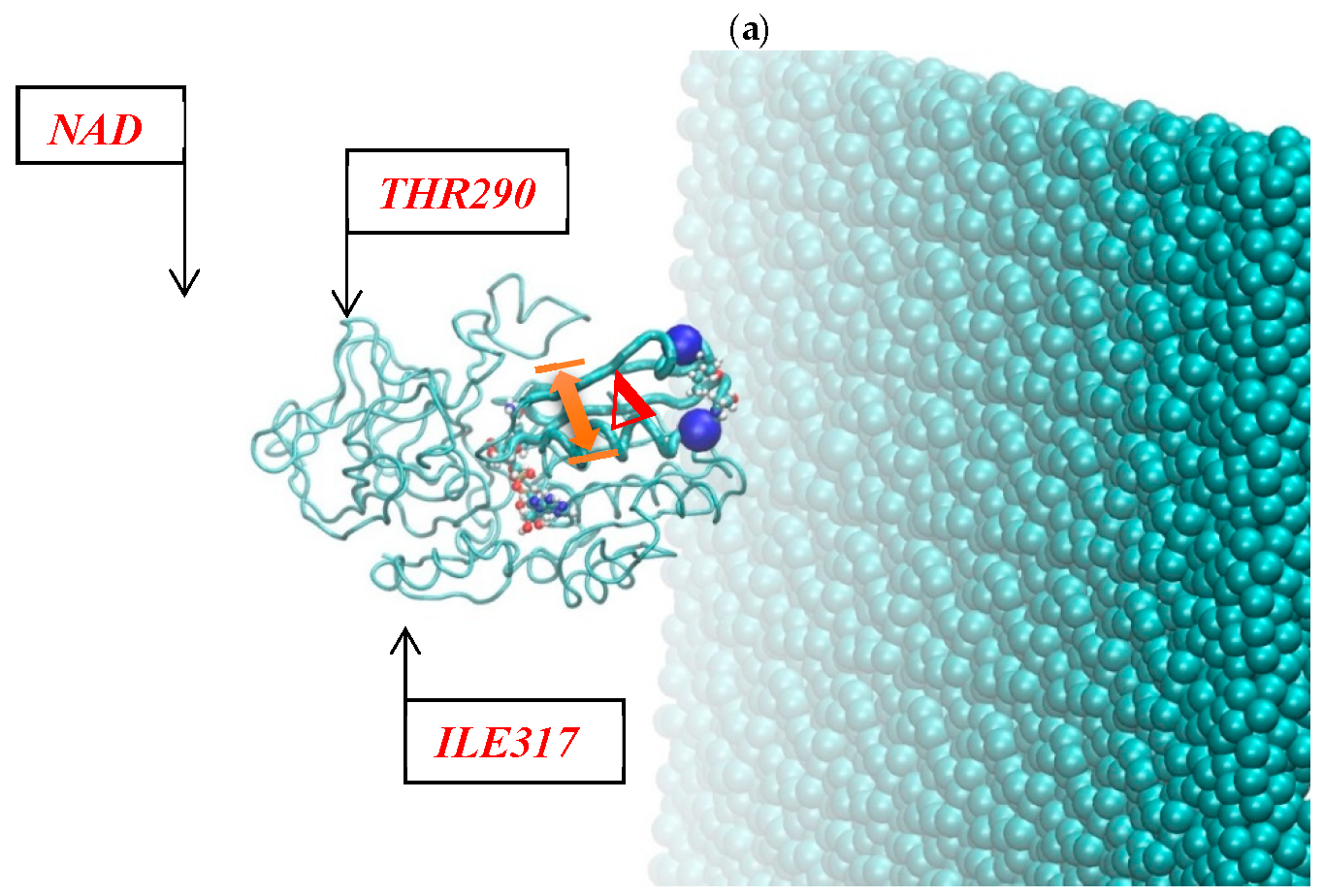
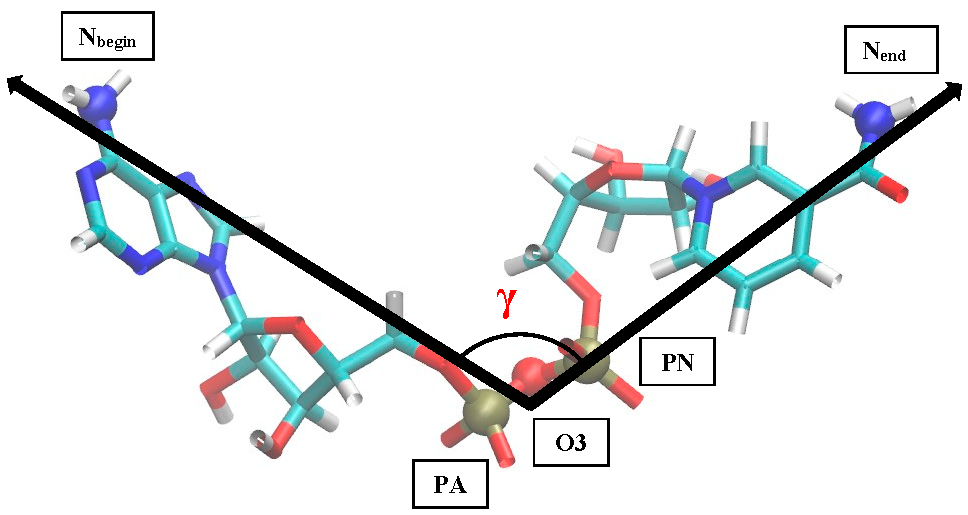
Figure 2 shows the resulting pictures of the adsorption of ADH+NAD on the surface of graphite carbon during prolonged 100 ns of dynamic changes from (a) the initially relaxed state to (b,c) intermediate states and (d) the final equilibrium condition. The enzyme ADH+NAD undergo multimillion conformational and rotational changes before adsorption on this graphite carbon (C-surface) in order to finally capture and relax on the surface. The dynamics of ADH+NAD adsorption on the graphite carbon surface was monitored using MD/AMBER calculations and Visual Molecular Dynamics (VMD) software. For each MD model, calculations were performed in 100 ns using the module “pmemd.cuda”. One of the nontrivial events of the conformational structural dynamics of the entire ADH+NAD+water system/C-surface and tracking of individual amino acid residues should be the behavior of the catalytic loops of the enzyme. Figure 2 show the dynamic patterns of the ADH+NAD/C-surface and the adsorption processes, accompanied by a gradual change in the orientation of the two catalytic loops of the ADH+NAD molecule relative to the graphite surface. An important observation is that these two catalytic loops are located close together inside the ADH+NAD molecule. However, upon reaching the adsorbing graphite surface, a separation between these loops is observed. A visual description of the entire process has shown in Figure 2, where two enlarged loops separate from each other and, consequently, open important catalytic ADH fragments as a result of the influence of the adsorbing C-surface of these two enzyme loops. Further, Figure 3 show the location of the above-mentioned two catalytic loops of the ADH+NAD molecule relative to the graphite surface. Two terminal amino acid residues THR290 and ILE317 are highlighted here, where Δ is introduced, as the distance between these two terminal amino acids. Figure 3a shows the location of the catalytic loops during adsorption of the ADH+NAD molecule on the carbon surface of graphite in the final (100 ns) state. Figure 3b shows the results of calculations of the dynamics of the distance between the catalytic loops of ADH+NAD on the carbon surface of graphite as a function of time. The results in Figure 3 clearly confirm the important observation mentioned above that the two catalytic loops separate from each other when they reach the adsorbing surface of graphite, whereas initially they were located close to each other inside the ADH+NAD molecule. The above observation and the presented data on the dynamics of the catalytic loops of ADH+NAD on the carbon surface correlate with the dynamical changes and rotations of the coenzyme NAD inside the ADH molecule. Using the data in Figure 3, the conformational changes of NAD during adsorption of ADH on the surface were also estimated, along with the open gap between the catalytic loops. In Figure 4, the positions of the atoms at the beginning and end of the chain NAD, as well as the atoms in the central region of NAD, are noted. Figure 4 demonstrate a method for estimating the angle of γ-vibration and rotation of coal in the three-dimensional structure of the NAD molecule, where auxiliary vectors are introduced and the starting points are designated A~ (PA, O3, PN), B~Nbegin, C~Nend with the results of calculating the dynamical changes in the angle of γ of the NAD molecule depending on time. They are shown in Figure 4b below. It can be seen that the dynamical changes of γ with time are completely correlated with the dynamics of the change in the distance between the catalytic loops of ADH + NAD on the carbon surface of graphite. The comparison of the results in Figure 3 and Figure 4 is straightforward. Thus, these results are the key features of the entire process of adsorption of ADH+NAD/C-surface, which is accompanied by a non-trivial structural transformation of the coenzyme NAD, correlating with the behavior of the catalytic loops of the enzyme ADH. Further, below in Figure 5, the orientations of the three titrated amino acid residues ASP54, HIS252 and ASP268 with time are shown. It is worth noting that for ADH+NAD+water+graphite surface, as described above, all calculations were performed at pH = 7 in a aqueous medium. The experimental data and analysis of the kinetic behavior of the ADH enzyme in solution as a function of pH and temperature, have shown that the highest activity is achieved in the pH range 7.0–8.0 and an optimal temperature about 303 K [4,5,6,7,26,27,28]. At pH 7, the protonation states of the catalytic triad, histidine (HIS), aspartate (ASP), and glutamate (GLU), are determined by their respective pKa values. Histidine has an imidazole side chain with a pKa of approximately 6.0, meaning that at physiological pH, it exists primarily in its deprotonated, neutral form. Aspartate (ASP) and glutamate (GLU), with pKa values of around 3.9 and 4.2, respectively, are both fully deprotonated at pH 7. In Figure 6, the dynamics of the angle of γ—internal vibration and rotation (N-CA-OD2) of three titrated amino acid residues ASP54 (a), HIS252 (b) and ASP268 (c) have been calculated. As in the case of the molecule above, auxiliary vectors were introduced, thereby thus, choosing the places of evaluation of amino acid residues in the form of: A~(CA)—the middle atom of the residue; B~Nbegin (NH3-end)—the residue of one terminal atom; C~OD2 (NE2)—the residue of another terminal atom. From the results shown in Figure 6, it can be seen that the dynamic behavior and the change in γ over time fully correlate with the dynamics of the change in the distance between the catalytic loops of ADH + NAD on the carbon surface of graphite, as shown in Figure 3b, as well as in Figure 4b. Figure 7 shows the location of the catalytic loops during adsorption of the ADH+NAD molecule on the carbon surface of graphite in the initial (a) state, (b) in the intermediate (50 ns) and (c) in the final (100 ns) states. Figure 8 demonstrates the procedure for calculating the estimate of the angle of γ-vibration and rotation of coal in the three-dimensional structure of the molecule NAD, where auxiliary vectors are introduced and the starting points are designated A~(PA,O3,PN), B~Nbegin, C~Nend. The dynamical changes in the angle γ of the NAD molecule as a function of time correlate with the dynamics of changes in the distance between the catalytic loops of ADH+NAD on the carbon surface of graphite: In Figure 8 below, the calculation of the dynamics of the NAD structure over time includes the angle between (point A~(PA, O3, PN), point B~Nbegin, point C~Nend). Thus, the orientation characteristics of the ADH enzyme’s titratable amino acid residues strongly correlate with the transformation of the NAD coenzyme. Thereby, the complete ADH+NAD structural behavior has to occur on the graphite surface during the adsorption processes. The ambient temperature and water environment during the implementation of the MD numerical experiment has been chosen. So far, the results in terms of pH and temperature coincide with data reported in the literature for the ADH enzyme in solution. Perhaps the key results, particularly Figure 3b and Figure 4b, which represent the main discoveries or observations, correlate well with experiments on the kinetics of the ADH enzyme. As can be seen from the MD results, presented in Figure 3b and Figure 4b, during the ADH+NAD adsorption the diffusion processes occur for 55 ns; after this period, ADH+NAD touch the graphite surface for a final relaxation on the surface. Following the adsorption of the ADH enzyme on the carbon surface, the coenzyme NAD from 55–60 ns begins to fluctuate, which is in full correlation with the opening of two catalytic loops associated with the NAD region. It is worth noting that current observation correlates with some experimental work related to the immobilization and adsorption of ADH enzyme on the carbon surface, the location of the enzyme and its fixation on carbon platforms. Moreover, the results also show that, in order to maintain an environment in which the ADH enzyme display good activity and to provide conditions for future technological applications, physiological conditions and ambient temperature can satisfactorily be applied to an enzymatic system involving the dehydrogenase enzyme. At the same time, the choice of carbon surfaces and platforms is motivated by a good control of the location of enzymes on the surface with very low enzyme consumption. The kinetic rates obtained for the ADH enzyme fixed to the carbon surface indicates a decrease in activity after the immobilization and fixation process. A significant loss of enzymatic activity is observed after immobilization, even though the affinity between the enzymes and their substrates and coenzymes persists. To date, the MD observations of the structural behavior of ADH+NAD, diffusion and adsorption processes correlate well with the kinetic rates obtained for an enzyme anchored by ADH on a carbon platform with various kinetic parameters (mainly the concentration of substrates and coenzyme), indicating the substantial changes (the loss or decrease) in the enzymatic activity of fixed enzymes after the immobilization process on the carbon surface. Judging by experiments and kinetic studies, this is due to a significant loss of enzymatic activity of ADH in the presence of a carbon surface, and the anchored enzyme should lose enzymatic activity. Thus, for the ADH enzyme and NAD coenzyme, the carbon platform has probably cause not only diffusion activity, but also some restriction in diffusion process, thereby making them difficult for the reduced form of the coenzyme to escape from the bulk solution (see, Supplementary Materials).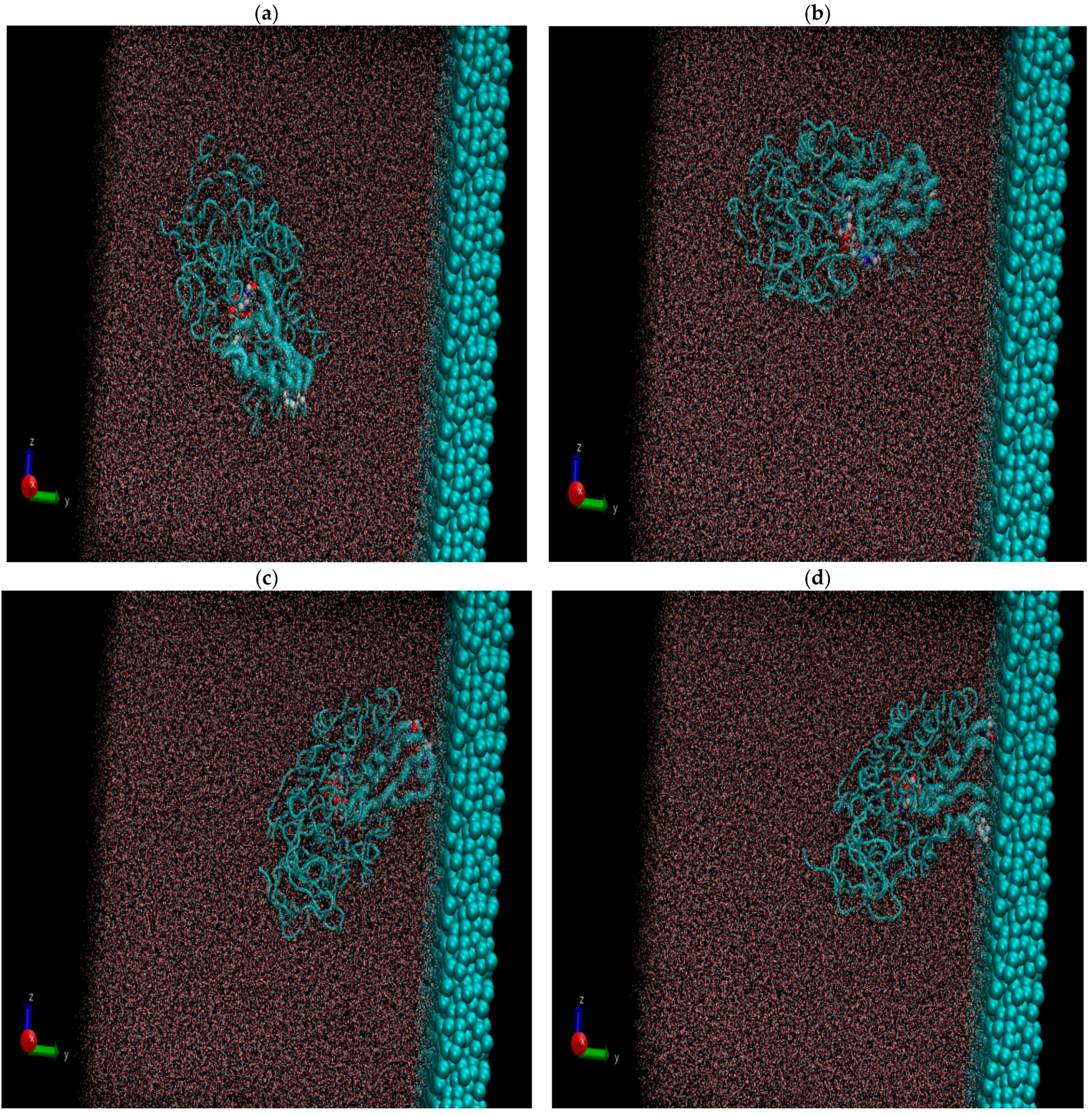
In conclusion, the structural conformations of the alcohol dehydrogenase (ADH) enzyme with its cofactor nicotinamide adenine dinucleotide (NAD) were studied on a graphite-like surface (C-surface) using the MD (molecular dynamics) modeling method. The enzyme ADH+NAD was modeled in an aqueous medium. The MD simulations, conducted using the AMBER-18 package (a fast implementation of the pmemd.cuda module), provide valuable statistical data on key structural aspects of ADH, particularly its conformational changes, including the rotation of the catalytic domain, the coenzyme binding domain, and rearrangements of the active center. This analysis is essential for understanding the atomic and molecular details of a catalytically active enzyme. The 100-ns dynamic simulations allowed to track the conformational and rotational changes of ADH+NAD in an aqueous environment. A non-trivial phenomenon has been observed: the conformational changes in ADH were correlated with structural transformations in its cofactor, NAD. However, it remains unclear whether the catalytic activity of ADH is preserved after such extensive conformational changes. Furthermore, the findings align with experimental studies on enzyme immobilization and adsorption onto carbon surfaces, as well as enzyme fixation on carbon platforms and oxide surfaces. The MD results of the distance and angle with time, appear to correlate well with experimental data on ADH+NAD kinetics, where the ADH+NAD diffusion process occurs over approximately 50–60 ns, after which the complex reaches the graphite surface and undergoes final relaxation. Following ADH adsorption onto the carbon surface, NAD begins to fluctuate between 55 and 60 ns, aligning with the opening of two catalytic loops associated with the NAD-binding region. Experimental and kinetic studies suggest that this structural change significantly impacts enzymatic activity, leading to the loss of catalytic function in the immobilized enzyme. For ADH and NAD species on carbon platforms, diffusion restrictions may arise, potentially preventing the reduced form of the coenzyme from flowing freely in the bulk solution. The ADH+NAD complex was preserved in the simulations, with the cofactor modeled within the enzyme structure as in the original PDB file. Interestingly, NAD can be efficiently stabilized outside the enzyme, maintaining its catalytic efficiency in a selected NAD-enriched solution, a specially designed buffer or solvent system to stabilize and preserve its activity.
| MDs | Molecular Dynamics Simulations |
| ADH | Alcohol Dehydrogenase |
| NAD | Nicotinamide Adenine Dinucleotide |
| VMD | Visual Molecular Dynamics |
| HIS | Histidine |
| ASP | Aspartate |
| GLU | Glutamate |
Conceptualization and supervision, P.P.G. and K.T.K.; methodology, K.T.K., M.A.I. and H.G.E.; validation, K.T.K., P.P.G. and I.A.B.; formal analysis, K.T.K., M.A.H. and I.A.B.; investigation, K.T.K., I.A.B. and M.A.H.; resources, K.T.K. and I.A.B.; data curation K.T.K., I.A.B. and P.P.G.; writing—original draft preparation, K.T.K., I.A.B. and P.P.G.; writing—review and editing, K.T.K., P.P.G., I.A.B., H.G.E. and M.A.I.; project administration, P.P.G. and K.T.K.; funding acquisition, P.P.G. and H.G.E. All authors have read and agreed to the published version of the manuscript.
Data supporting the results of this study are available upon request from the corresponding author.
Not applicable.
The authors declare no conflicts of interest regarding this manuscript.
This work was supported by funding from the JINR—ASRT (ARE) grant “Analysis of molecular modeling of the effect of nanometallic oxides on biological molecules”. The work was carried out with the support of the Ministry of Science and Education of the Republic of Tajikistan within the framework of the project: “Molecular orientation of DNA on biocompatible metal-oxide materials” (Grant No.0122TJ1327).
The work has been performed within the framework of the joint research program of the JINR—Dubna State University. The work was performed within the framework of the state assignment of the Ministry of Science and Higher Education of the Russian Federation (No. 1024011000011-7-1.4.2;3.5.2 Conjugates boron-containing quantum dots with biovectors for the diagnosis and boron-neutron capture therapy of superficial malignant tumors (FEEM-2024-0011)). The MD calculations were performed on the servers of the Heterogeneous HybriLIT platform of the Multifunctional Information and Computing Complex (IVC), MLIT (Laboratory of Information Technology named after M.G. Meshcheryakov), JINR (Joint Institute for Nuclear Research).
[1] P.S. Wagenknecht, J.M. Penney, R.T. Hembre, "Transition Metal-Catalyzed Regeneration of Nicotinamide Coenzymes" Organometallics, vol. 22, pp. 1180-1182, 2003. [Crossref]
[2] K. Nakamura, R. Yamanaka, "Light Mediated Cofactor Recycling System in Biocatalytic Asymmetric Reduction of Ketenes" Chem. Commun., vol. 16, pp. 1782-1783, 2002. [Crossref] [PubMed]
[3] D.S. Bilan, V.V. Belousov, "Genetically Encoded Probes for NAD+/NADH Monitoring" Free Radic. Biol. Med., vol. 100, pp. 32-42, 2016. [Crossref] [PubMed]
[4] A.V. Presečki, Đ. Vasić-Rački, "Modelling of the Alcohol Dehydrogenase Production in Baker’s Yeast" Process Biochem., vol. 40, pp. 2781-2791, 2005. [Crossref]
[5] A. Tušek, A. Šalić, Ž. Kurtanjek, B. Zelić, "Modelling and Kinetic Parameter Estimation of Alcohol Dehydrogenase Catalyzed Hexanol Oxidation in a Microreactor" Eng. Life Sci., vol. 12, pp. 49-56, 2012. [Crossref]
[6] B. Orlich, H. Berger, M. Lade, R. Schomäcker, "Stability and Activity of Alcohol Dehydrogenases in W/O-Microemulsions: Enantioselective Reduction Including Cofactor Regeneration" Biotechnol. Bioeng., vol. 70, pp. 638-646, 2001. [PubMed]
[7] S. Höhn, K. Zheng, S. Romeis, M. Brehl, W. Peukert, D. de Ligny, et al., "Effects of Medium pH and Preconditioning Treatment on Protein Adsorption on 45S5 Bioactive Glass Surfaces" Adv. Mater. Interfaces, vol. 7, p. 2000420, 2020. [Crossref]
[8] T.E. Benavidez, D. Torrente, M. Marucho, C.D. Garcia, "Adsorption and Catalytic Activity of Glucose Oxidase Accumulated on OTCE upon the Application of External Potential" J. Colloid Interface Sci., vol. 435, pp. 164-170, 2014. [Crossref] [PubMed]
[9] F. Wang, Y.-Q. Zhang, "Protein and Peptide Nanoparticles for Drug Delivery" Adv. Protein Chem. Struct. Biol., vol. 98, pp. 263-291, 2015. Available online: https://www.sciencedirect.com/bookseries/advances-in-protein-chemistry-and-structural-biology/vol/98/suppl/C.
[10] V.V. Welborn, "Structural Dynamics and Computational Design of Synthetic Enzymes" Chem. Catal., vol. 2, pp. 19-28, 2022. [Crossref]
[11] W. Norde, J. Lyklema, "Why Proteins Prefer Interfaces" J. Biomater. Sci. Polym. Ed., vol. 2, pp. 183-202, 1991. [Crossref] [PubMed]
[12] J.D. Andrade, "," in Surface and Interfacial Aspects of Biomedical Polymers: Volume 1 Surface Chemistry and Physics, , Eds. Berlin, Germany: Springer Science & Business Media, 2012, .
[13] P.P. Gladyshev, A. Shapovalov Yu, V.P. Kvasova, "," in Reconstructed Oxidoreductase Systems, , Eds. KazSSR: NAUKA (The Science), 1987, . (In Russian)
[14] P.P. Gladyshev, M.I. Goryaev, I.G. Shpilberg, Y.A. Shapovalov, "Sorption Immobilization of NAD-Dependent Enzyme Systems. I. The Influence of Electrostatic Interactions on the Orientation of Alcohol Dehydrogenase on the Surface of the Carrier" Mol. Biol., vol. 16, pp. 938-942, 1982. (In Russian) [PubMed]
[15] P.P. Gladyshev, M.I. Goryaev, I.G. Shpilberg, "Sorption Immobilization of NAD-Dependent Enzyme Systems. II. The Influence of Hydrophobic Interactions on the Orientation of Alcohol Dehydrogenase on the Surface of the Carrier" Mol. Biol., vol. 16, pp. 943-947, 1982. (In Russian) [PubMed]
[16] J. Foresman, E. Frish, "," in Exploring Chemistry, , Eds. Pittsburg, PA, USA: Gaussian Inc., 1996, . Available online: https://gaussian.com/expchem3/.
[17] A.R. Leach, "," in Molecular Modelling: Principles and Applications, , Eds. Hoboken, NJ, USA: Pearson Education, 2001, . Available online: http://inis.jinr.ru/sl/Simulation/Leach%20A.,%20Molecular%20Modelling.%20Principles%20and%20Applications,%202001.pdf.
[18] D.A. Case, T.E. Cheatham, T. Darden, H. Gohlke, R. Luo, K.M. Merz, et al., "The Amber Biomolecular Simulation Programs" J. Computat. Chem., vol. 26, pp. 1668-1688, 2005. [Crossref] [PubMed]
[19] D.A. Case, H.M. Aktulga, K. Belfon, D.S. Cerutti, G.A. Cisneros, V.W.D. Cruzeiro, et al., "AmberTools" J. Chem. Inf. Model., vol. 63, pp. 6183-6191, 2023. [Crossref]
[20] T.-S. Lee, D. Mermelstein, C. Lin, S. LeGrand, T.J. Giese, A. Roitberg, et al., "GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features" J. Chem. Inf. Mod., vol. 58, pp. 2043-2050, 2018. [Crossref]
[21] V.W.D. Cruzeiro, M.S. Amaral, A.E. Roitberg, "Redox Potential Replica Exchange Molecular Dynamics at Constant pH in AMBER: Implementation and Validation" J. Chem. Phys., vol. 149, p. 072338, 2018. [Crossref]
[22] K. Kholmurodov, "," in Models in Bioscience and Materials Research: Molecular Dynamics and Related Techniques, , Eds. Hauppauge, NY, USA: Nova Science Publishers Ltd., 2013, . Available online: https://novapublishers.com/shop/models-in-bioscience-and-materials-research-molecular-dynamics-and-related-techniques/.
[23] K. Kholmurodov, "," in Computational Materials and Biological Sceinces, , Eds. Hauppauge, NY, USA: Nova Science Publishers Ltd., 2015, .
[24] K.T. Kholmurodov, I.A. Baigunov, P.P. Gladyshev, H. Elhaes, M. Ibrahim, "The Molecular Dynamics and Experimental Studies of the Structural Behavior of Alcoholdehydrogenase Enzyme on the Graphitic Sorbent Surfaces" Res. Dev. Mater. Sci., vol. 20, p. RDMS.000982, 2024. [Crossref]
[25] S.A. Neto, J.C. Forti, V. Zucolotto, P. Ciancaglini, A.R. De Andradea, "The Kinetic Behavior of Dehydrogenase Enzymes in Solution and Immobilized onto Nanostructured Carbon Platforms" Process Biochem., vol. 46, pp. 2347-2352, 2011. [Crossref]
[26] J.C. Forti, S.A. Neto, V. Zucolotto, P. Ciancaglini, A.R. De Andrade, "Development of Novel Bioanodes for Ethanol Biofuel Cell Using PAMAM Dendrimers as Matrix for Enzyme Immobilization" Biosens. Bioelectron., vol. 26, pp. 2675-2679, 2011. [Crossref]
[27] D. Sokic-Lazic, S.D. Minteer, "Citric Acid Cycle Biomimic on a Carbon Electrode" Biosens. Bioelectron., vol. 24, pp. 939-944, 2008. [Crossref] [PubMed]
[28] M. Radović, L. Hok, M. Panić, M. Cvjetko Bubalo, R. Vianello, M. Vinković, et al., "Deep Eutectic Solvents as a Stabilising Medium for NAD Coenzyme: Unravelling the Mechanism behind Coenzyme Stabilisation Effect" Green Chem., vol. 24, pp. 7661-7674, 2022. [Crossref]
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more